For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPMSTLDQIDGLIRIETSPKEKATPVIMEMMRSVYPHDDVFGTYCTVNEYVDCPPDELFDYMSDTRCLEE WTYSLRGFTPTDEPDLWLAHDRLGAGPDGPGSAIYTRTVANREALTVDYHCAWDQGKHLWMIYLMRIVDA QVVFDKPGSVVLWTNCHHPFYDRNPYPETAPAGRPVWVGDFWDMFGPGHLLELRNLKAIAEYRHRNGLPV TPNWMR
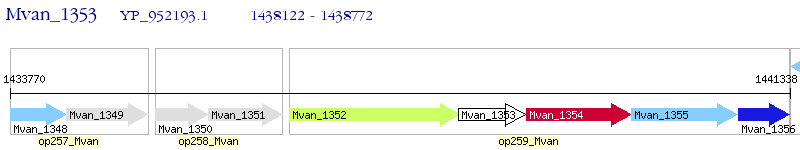
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1353 | - | - | 100% (216) | hypothetical protein Mvan_1353 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2166 | - | 3e-99 | 75.47% (212) | hypothetical protein MAB_2166 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_4423 | - | 1e-108 | 81.90% (210) | hypothetical protein MAV_4423 |
| M. smegmatis MC2 155 | MSMEG_1489 | - | 1e-115 | 85.85% (212) | hypothetical protein MSMEG_1489 |
| M. thermoresistible (build 8) | TH_1539 | - | 1e-103 | 78.95% (209) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2166|M.abscessus_ATCC_1997 MTDTIAKTANPGLKALPALSDIAAEAGTEDALPGVHRIETSPREKATPII
MAV_4423|M.avium_104 -------------MSLPALEDIG------DLIDGVTRIETSPREKATPII
TH_1539|M.thermoresistible__bu -------------MSLPALEDIAVHRGSTEPIDGVIRIETSPREKATPII
Mvan_1353|M.vanbaalenii_PYR-1 -------------MPMS----------TLDQIDGLIRIETSPKEKATPVI
MSMEG_1489|M.smegmatis_MC2_155 -------------MPLPALEDITIHRGTADPIDGLIRIETSPKEAATPII
.:. : : *: ******:* ***:*
MAB_2166|M.abscessus_ATCC_1997 MDMMRSVYPHDQVFGPYCTVQEYVDCPPDDLHRYLSDTRCLEEWTYSLRG
MAV_4423|M.avium_104 MDMMRSVYPHDQVFGQYCTVNDYIDCPPDDLFEYMADTRSLEEWTYSLRG
TH_1539|M.thermoresistible__bu MDMIRSVYPHDDVYGPFCTVQDYVDCPPDELYDWMSDTRCLEEWTYSLRG
Mvan_1353|M.vanbaalenii_PYR-1 MEMMRSVYPHDDVFGTYCTVNEYVDCPPDELFDYMSDTRCLEEWTYSLRG
MSMEG_1489|M.smegmatis_MC2_155 MEMMRSVYPHEQVFGEYCTVNDYIACPPDELFDYLADTRSLEEWTYSLRG
*:*:******::*:* :***::*: ****:*. :::***.**********
MAB_2166|M.abscessus_ATCC_1997 FAETAEPGLWVAHDRLG-----SDTLIYTRTESNAEAGTVDYHCAWDQGD
MAV_4423|M.avium_104 FTPTDEPGLWLAHDRLG-----SETKIYTRTVANAQARTVDYHCAWDQGD
TH_1539|M.thermoresistible__bu FEETDEPGLWLAWDRLG-----SETKIYTRTVANPQARIVDYHCAWDQGK
Mvan_1353|M.vanbaalenii_PYR-1 FTPTDEPDLWLAHDRLGAGPDGPGSAIYTRTVANREALTVDYHCAWDQGK
MSMEG_1489|M.smegmatis_MC2_155 FATTEEPGLWVARDRLGEGSGGPGSAIYTRTVANRDAMTVDYHCAWDQGK
* * **.**:* **** . : ***** :* :* **********.
MAB_2166|M.abscessus_ATCC_1997 HLWMIYLMRVVDAQVVLNKPGSVVLWTNCHHPFYDENPYPETAPPQRKPW
MAV_4423|M.avium_104 HLWMIYLMRVVDAQTVLDKPGSVVLWTNCHHPFYDHNPYPQTAPPQRPVW
TH_1539|M.thermoresistible__bu HLWMIYLMRVIDAQLVLDKPGSVVLWTNCHHPFYDENPYPETAPPERPVW
Mvan_1353|M.vanbaalenii_PYR-1 HLWMIYLMRIVDAQVVFDKPGSVVLWTNCHHPFYDRNPYPETAPAGRPVW
MSMEG_1489|M.smegmatis_MC2_155 HLWMIYLMRIVDAQVVFDKPGSVVLWTNCHHPFYDENPYPETAPPGRPVW
*********::*** *::*****************.****:***. * *
MAB_2166|M.abscessus_ATCC_1997 VGDFWDMFGAGHMLELLNLKAIAEYRHSHGLPIVPEWMK
MAV_4423|M.avium_104 VGDFWDMFGAGHLLELRNLKAIAEYRHRHGLPVTPVWMR
TH_1539|M.thermoresistible__bu VGDFWDMFGAGHALELRNLKRIAEYRHHNGLPVKPLWMS
Mvan_1353|M.vanbaalenii_PYR-1 VGDFWDMFGPGHLLELRNLKAIAEYRHRNGLPVTPNWMR
MSMEG_1489|M.smegmatis_MC2_155 VGDFWDMFGPGHLLELKNLKAIAEYRHRNGLPLTPAWMK
*********.** *** *** ****** :***: * **