For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAKADKATAVAEIAEKFKEATAAVVTEYRGLTVSNLAELRRSLGNSTTYTVAKNTLVKRAAVEAGVEGLD DMFAGPTAIAFINGEPVDAAKAIKKFAKDNKALIVKGGYMDGRALTVGEVERIADLESREVLLSKLAGAM KAKQSQAAALFVAPASQVARLAAALQEKKAAGDSAESAA
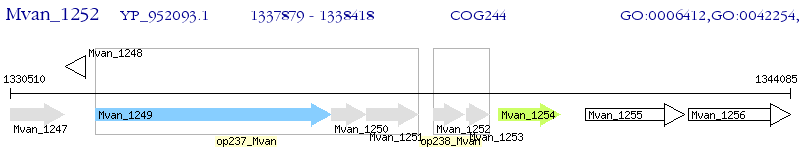
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1252 | rplJ | - | 100% (179) | 50S ribosomal protein L10 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0670 | rplJ | 9e-71 | 76.40% (178) | 50S ribosomal protein L10 |
| M. gilvum PYR-GCK | Mflv_5101 | rplJ | 1e-85 | 90.50% (179) | 50S ribosomal protein L10 |
| M. tuberculosis H37Rv | Rv0651 | rplJ | 1e-70 | 76.40% (178) | 50S ribosomal protein L10 |
| M. leprae Br4923 | MLBr_01896 | rplJ | 9e-69 | 75.88% (170) | 50S ribosomal protein L10 |
| M. abscessus ATCC 19977 | MAB_3877c | rplJ | 2e-73 | 80.57% (175) | 50S ribosomal protein L10 |
| M. marinum M | MMAR_0990 | rplJ | 9e-73 | 77.65% (179) | 50S ribosomal protein L10, RplJ |
| M. avium 104 | MAV_4508 | rplJ | 8e-74 | 79.10% (177) | 50S ribosomal protein L10 |
| M. smegmatis MC2 155 | MSMEG_1364 | rplJ | 2e-74 | 80.57% (175) | 50S ribosomal protein L10 |
| M. thermoresistible (build 8) | TH_1855 | rplJ | 5e-73 | 78.29% (175) | PROBABLE 50S RIBOSOMAL PROTEIN L10 RPLJ |
| M. ulcerans Agy99 | MUL_0741 | rplJ | 6e-73 | 77.65% (179) | 50S ribosomal protein L10 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1364|M.smegmatis_MC2_155 MAKADKATAVADIAEQFKASTATVVTEYRGLTVANLAELRRALGDSATYT
TH_1855|M.thermoresistible__bu MARADKAAAVAEIAERFKEATATVVTEYRGLTTADLAELRRALGESASYR
Mb0670|M.bovis_AF2122/97 MARADKATAVADIAAQFKESTATLITEYRGLTVANLAELRRSLTGSATYA
Rv0651|M.tuberculosis_H37Rv MARADKATAVADIAAQFKESTATLITEYRGLTVANLAELRRSLTGSATYA
MLBr_01896|M.leprae_Br4923 MARADKATAVANIAEQFKAATAALITEYRGLTVANLAELRRSLTGSASYA
MAV_4508|M.avium_104 MARADKATAVADIAEQFKEATATLITEYRGLTVANLAELRRSLSGAATYS
MMAR_0990|M.marinum_M MARADKATAVADIAEHFKASTATVITEYRGLTVSNLAELRRSLGESATYA
MUL_0741|M.ulcerans_Agy99 MARADKATAVADIAEHFKASTATVITEYRGLTVSNLAELRRSLGESATYA
MAB_3877c|M.abscessus_ATCC_199 MAKTDKVTAVAEITEQFKDSTATVITEYRGLSVSALATLRRSLGASATYA
Mvan_1252|M.vanbaalenii_PYR-1 MAKADKATAVAEIAEKFKEATAAVVTEYRGLTVSNLAELRRSLGNSTTYT
Mflv_5101|M.gilvum_PYR-GCK MAKADKATAVADIAEKFKESTATVVTEYRGLTVSNLAELRRSLGSSTTYT
**::**.:***:*: :** :**:::******:.: ** ***:* :::*
MSMEG_1364|M.smegmatis_MC2_155 VAKNTLVKRAASEAGIEGLDELFAGPTAIAFVKGEAVDAAKAIKKFAKDN
TH_1855|M.thermoresistible__bu VAKNTLVQRAAEEAGIEGLDELFSGPTAIAFVTGEPVDAAKAIKKFAKDN
Mb0670|M.bovis_AF2122/97 VAKNTLIKRAASEAGIEGLDELFVGPTAIAFVTGEPVDAAKAIKTFAKEH
Rv0651|M.tuberculosis_H37Rv VAKNTLIKRAASEAGIEGLDELFVGPTAIAFVTGEPVDAAKAIKTFAKEH
MLBr_01896|M.leprae_Br4923 VAKNTLIKRAASEVGIEGLDELFAGPTAIAFVTGEPVDAAKAIKTFAKEH
MAV_4508|M.avium_104 VAKNTLVKRAASEAGIEGLDELFAGPTAIAFVTGEPVDAAKAIKTFAKEH
MMAR_0990|M.marinum_M VAKNTLIKRAASEAGVEGLDELFAGPTAIAFVKGEPVDAAKALKTFAKEH
MUL_0741|M.ulcerans_Agy99 VAKNTLIKRAASEAGVEGLDELFAGPTAIAFVKGEPVDAAKALKTFAKEH
MAB_3877c|M.abscessus_ATCC_199 VAKNTLVKRAAADAGVEGLDELFAGPTAIAFIKGEPVDAAKAIKTFAKDN
Mvan_1252|M.vanbaalenii_PYR-1 VAKNTLVKRAAVEAGVEGLDDMFAGPTAIAFINGEPVDAAKAIKKFAKDN
Mflv_5101|M.gilvum_PYR-GCK VAKNTLVKRAAAEAGIEGLDELFVGPTAIAFIEGEPVDAAKALKKFAKDN
******::*** :.*:****::* *******: **.******:*.***::
MSMEG_1364|M.smegmatis_MC2_155 KALVIKGGYMDGKALSVADVEKIADLESREVLLAKLAGAMKGNLSKAAGL
TH_1855|M.thermoresistible__bu KALVIKGGYMDGRPLSVAEIEKIADLESREVLLAKLAGAMKGNLAKAAGL
Mb0670|M.bovis_AF2122/97 KALVIKGGYMDGHPLTVAEVERIADLESREVLLAKLAGAMKGNLAKAAGL
Rv0651|M.tuberculosis_H37Rv KALVIKGGYMDGHPLTVAEVERIADLESREVLLAKLAGAMKGNLAKAAGL
MLBr_01896|M.leprae_Br4923 KALVIKGGYMDGHPMTVAEVERIADLESREVLLAKLAGAMKGTFAKAIGL
MAV_4508|M.avium_104 KALVIKGGYMDGRALSVAEVERIADLESREVLLAKLAGAMKGNLAKAAGL
MMAR_0990|M.marinum_M KALVIKGGYMDGHALTVAEVERIADLETREVLLAKLAGAMKGNLAKAAGL
MUL_0741|M.ulcerans_Agy99 KALVIKGGYMDGHALTVAEVERIADLETREVLLAKLAGAMKGNLAKAAGL
MAB_3877c|M.abscessus_ATCC_199 KALIIKGGYMDGRALSVAEVERIADLESREVLLAKLAGAMKGNLNKAAGL
Mvan_1252|M.vanbaalenii_PYR-1 KALIVKGGYMDGRALTVGEVERIADLESREVLLSKLAGAMKAKQSQAAAL
Mflv_5101|M.gilvum_PYR-GCK KALVVKGGYMDGRALSVSEVERIADLESREVLLSKFAGALKAKQSQAAAL
***::*******:.::*.::*:*****:*****:*:***:*.. :* .*
MSMEG_1364|M.smegmatis_MC2_155 FNAPASQVARLAAALQEKKAGE-----EAA--------------------
TH_1855|M.thermoresistible__bu FNAPASQVARLAAALQEKKPAE-----DAA--------------------
Mb0670|M.bovis_AF2122/97 FNAPASQLARLAAALQEKKACP---GPDSAE-------------------
Rv0651|M.tuberculosis_H37Rv FNAPASQLARLAAALQEKKACP---GPDSAE-------------------
MLBr_01896|M.leprae_Br4923 FNAPTSQMARLTAALQEKKAF----EPASAE-------------------
MAV_4508|M.avium_104 FNAPASQVARLAAALQEKKAAE---CPAEAPQPAAET-PAEAPEAPADAE
MMAR_0990|M.marinum_M FNAPASQFARLAAALQEKKAAEPGSAPAAATETPEAAGAAEASEAPEATD
MUL_0741|M.ulcerans_Agy99 FNAPASQFARLAAALQEKKAAEPGSAPAAATETPEAAGAAEASEAPEATD
MAB_3877c|M.abscessus_ATCC_199 FAAPASQVARLAAALQEKKAGE-----ESAA-------------------
Mvan_1252|M.vanbaalenii_PYR-1 FVAPASQVARLAAALQEKKAAG-----DSAESAA----------------
Mflv_5101|M.gilvum_PYR-GCK FVAPASQVARLAAALQEKKSAG-----DSAESAA----------------
* **:**.***:*******. *
MSMEG_1364|M.smegmatis_MC2_155 ------
TH_1855|M.thermoresistible__bu ------
Mb0670|M.bovis_AF2122/97 ------
Rv0651|M.tuberculosis_H37Rv ------
MLBr_01896|M.leprae_Br4923 ------
MAV_4508|M.avium_104 ------
MMAR_0990|M.marinum_M TATDAE
MUL_0741|M.ulcerans_Agy99 TATDAE
MAB_3877c|M.abscessus_ATCC_199 ------
Mvan_1252|M.vanbaalenii_PYR-1 ------
Mflv_5101|M.gilvum_PYR-GCK ------