For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RDRSHLRVVTDLESRPDLRHPRRSDVVVYRVRVDIDDADPPIWRRIDLRSDVTLDFVHQVLQVAFDWTDS HLHRFSLGGGAFDRDSQLFLCPYDVAEADVEDDGVPAAGTRLDETLTGPGDVLHYLYDYGDNWELTLRLE QVLPADGDCAAAVVVDGERAAPPEDCGHLVDAASLAEVLPDPSLLELGRINRSLRGPYFVLREAKVDPRL IDLVHQLSYTEHGDSLAEMALTLVAEPATPDDDDLIRHLRAFQWFLDRAADGGIALTAAGYMKPNDVVEA SKVVPVMADWIGTKNREAHCYPLLQFRETVQDLGLLRKNKGQLLLTKAGQSAQRDVNALWTHLASRLNPK PGDEFHVQAGLVMLAYAAASPNAELPLNTISAILTELGWRHANGTPVTDSALRHLPVYEVLTNVTDQPVT WRRRDLISPVAARLARKALEV*
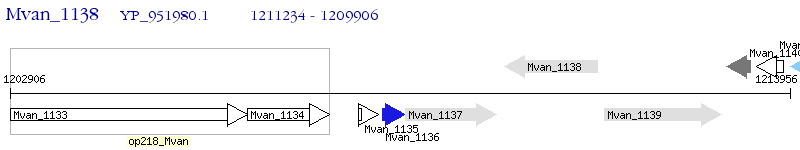
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1138 | - | - | 100% (442) | plasmid pRiA4b ORF-3 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5199 | - | 0.0 | 84.32% (440) | plasmid pRiA4b ORF-3 family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6698 | - | 7e-29 | 46.62% (148) | IS1096, tnpR protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1138|M.vanbaalenii_PYR-1 -----------------------------MRDRSHLRVVTDLESRPDLRH
Mflv_5199|M.gilvum_PYR-GCK MVPGEHTHPRAARLYSDQDELDESCDASPMRDRSHLRVVTDLASRPDLRH
MSMEG_6698|M.smegmatis_MC2_155 ---------------------------------MFDEPTPEVRAVPDRAR
. . ..:: : ** :
Mvan_1138|M.vanbaalenii_PYR-1 PRRSDVVVYRVRVDIDDADPPIWRRIDLRSDVTLDFVHQVLQVAFDWTDS
Mflv_5199|M.gilvum_PYR-GCK PRRSDVVVYRVRVDIDDADPPIWRRIDLRSDLTLDVVHQVLQVAFDWTDS
MSMEG_6698|M.smegmatis_MC2_155 G-------FRVRVDLMYAKPPIWRRLDLPGDLMLDELHVVLQVVMGWQDS
:*****: *.******:** .*: ** :* ****.:.* **
Mvan_1138|M.vanbaalenii_PYR-1 HLHRFSLGGGAFDRDSQLFLCPYDVAEADVEDDGVPAAGTRLDETLTGPG
Mflv_5199|M.gilvum_PYR-GCK HLHRFSLGGGAFDWDSQLFLCPYDVVEPDFEDDGVPAAETHLDETLTATG
MSMEG_6698|M.smegmatis_MC2_155 HLHKFGVG--ADRRTRAYFVTGFDLSEG---DDGVVEDSVRLDQVVSDKG
***:*.:* * *: :*: * **** .:**:.:: *
Mvan_1138|M.vanbaalenii_PYR-1 DVLHYLYDYGDNWELTLRLEQVLPADGDCAAAVVVDGERAAPPEDCGHLV
Mflv_5199|M.gilvum_PYR-GCK DALHYLYDYGDNWELTLRLEQVLPAEGACAAAVVVDGERAAPPEDCGHLV
MSMEG_6698|M.smegmatis_MC2_155 ERLFYDYDFGDGWDHVLVVEDVFDDP--PPAAVCLTGKMACPPEDCGGLG
: *.* **:**.*: .* :*:*: .*** : *: *.****** *
Mvan_1138|M.vanbaalenii_PYR-1 DAASLAEVLPDPSLLELGRINRSLRGPYFVLREAKVDPRLIDLVHQLSYT
Mflv_5199|M.gilvum_PYR-GCK EAASLAEVLPDPARFEQDRINDALRGPFFVLREAKVDPRLVDLVHRLGYT
MSMEG_6698|M.smegmatis_MC2_155 GYEELAAWVRGG----------------YDPRETPMGLGAQEMRDWLPRG
.** : . : **: :. :: . *
Mvan_1138|M.vanbaalenii_PYR-1 EHGDSLAEMALTLVAEPATPDDDDLIRHLRAFQWFLDRAADGGIALTAAG
Mflv_5199|M.gilvum_PYR-GCK AHGDSLSHAALALTAEPARLEAADLVGHLRGFQWFLDRAADGGIPLTGAG
MSMEG_6698|M.smegmatis_MC2_155 WHPDRFSVAETNDALAALNTR-----------------------------
* * :: . .
Mvan_1138|M.vanbaalenii_PYR-1 YMKPNDVVEASKVVPVMADWIGTKNREAHCYPLLQFRETVQDLGLLRKNK
Mflv_5199|M.gilvum_PYR-GCK YLKPADVVEASKVVPVMADWIGAKNREAHCYPLLQFREMLQDLGLLRKYK
MSMEG_6698|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1138|M.vanbaalenii_PYR-1 GQLLLTKAGQSAQRDVNALWTHLASRLNPKPGDEFHVQAGLVMLAYAAAS
Mflv_5199|M.gilvum_PYR-GCK GQLLLTKAGQAAQRDVNALFEHLACRLNPKPGDEFHVQASLLTLTYAAAS
MSMEG_6698|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1138|M.vanbaalenii_PYR-1 PNAELPLNTISAILTELGWRHANGTPVTDSALRHLPVYEVLTNVTDQPVT
Mflv_5199|M.gilvum_PYR-GCK PDAQLPLSTIAAILTELGWRHANGIPVTESALRHLPVYEVLSNVTDQPAT
MSMEG_6698|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1138|M.vanbaalenii_PYR-1 WRRRDLISPVAARLARKALEV
Mflv_5199|M.gilvum_PYR-GCK WRRRNVISPGAAALARRALGL
MSMEG_6698|M.smegmatis_MC2_155 ---------------------