For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ASRGRYSQPSDGAVFCAFLLLLLAVGFVIKYIWWFVGGAVLVGVFAGIYWLAQKMEEERQLEADEAAERE FEMTRRAERQKRWTLIGDTRAIYGEDGAAAMRKITDDPEIDAEAAPAQDDSPFAQLATTSAELDALVREK PSAWPHALFASILVQRAAPLQSRLRDSELGFTAPTSTVALTPVHLASTVVTLIDEMIATAHQLEGFMAAP AFMAAFAGKDGEEADPEAIKHIAHRTMDYLERFMELSERCRALPVRSEHVDIVADCARMLDDPLASYREF IADFVDIIEALPRVLTHATGPVDMGSLGLYLKIDDKRHSRVLTRLDAITKS*
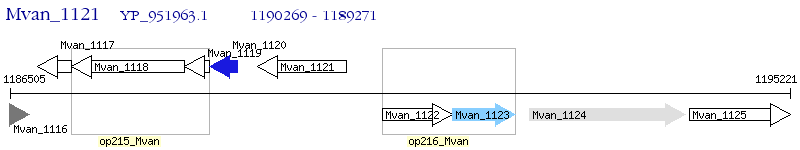
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1121 | - | - | 100% (332) | hypothetical protein Mvan_1121 |
| M. vanbaalenii PYR-1 | Mvan_4903 | - | 2e-20 | 28.64% (206) | hypothetical protein Mvan_4903 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5178 | - | 7e-98 | 56.88% (327) | hypothetical protein Mflv_5178 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1270 | - | 2e-12 | 36.78% (87) | hypothetical protein MSMEG_1270 |
| M. thermoresistible (build 8) | TH_2742 | - | 4e-33 | 41.62% (173) | PUTATIVE putative hypothetical protein Mflv_5178 |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1121|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_5178|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1270|M.smegmatis_MC2_155 --------------------------------------------------
TH_2742|M.thermoresistible__bu VDRESEPSRSRAGRVGAKAFTAGVTVATAGALAVAPVVVTATPPDVRVEP
Mvan_1121|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_5178|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1270|M.smegmatis_MC2_155 --------------------------------------------------
TH_2742|M.thermoresistible__bu SVADVQLSAFQNPLEVWLDVFTGPTGLLYQLETLGERTSTHWRALADALG
Mvan_1121|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_5178|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1270|M.smegmatis_MC2_155 --------------------------------------------------
TH_2742|M.thermoresistible__bu DPEVQARFTSFIRNTIDDPMRIPNALLAAPGKYGPVLSAAAQNTGQALVE
Mvan_1121|M.vanbaalenii_PYR-1 ---------------------------MASRGRYSQPSDGAVFCAFLLLL
Mflv_5178|M.gilvum_PYR-GCK ---------------------------MAD---HSSPGDGSVLVAIFVLL
MSMEG_1270|M.smegmatis_MC2_155 ------------------------------------------------MV
TH_2742|M.thermoresistible__bu AFAGLFFERQARTERGALLWLDENGQPTTDETITGIPHMAPPLIQETIRL
:
Mvan_1121|M.vanbaalenii_PYR-1 LAVG-----FVIKYIWWFVG-----------------------GAVLVGV
Mflv_5178|M.gilvum_PYR-GCK IIAG-----LVVRYIWWVVG-----------------------AAALAGV
MSMEG_1270|M.smegmatis_MC2_155 VVIG-----FIIKYIWWILG-----------------------GLAMVAA
TH_2742|M.thermoresistible__bu LAEGNFTDAFAEVNLWFLMDVLSDQRAGFLDALRVPGNFLEDLGAETLAR
: * : :*:.:. . .
Mvan_1121|M.vanbaalenii_PYR-1 FAGIYWLAQKMEEERQLE--------------------------------
Mflv_5178|M.gilvum_PYR-GCK VYLCVVLTRKAEERRLRD--------------------------------
MSMEG_1270|M.smegmatis_MC2_155 FFIIRALVR----WHLAA--------------------------------
TH_2742|M.thermoresistible__bu VLGTTWMDPLDEDGEIRHGWGPGLLSRARLGNFARAILAPPVTALFQTME
. : .
Mvan_1121|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_5178|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1270|M.smegmatis_MC2_155 --------------------------------------------------
TH_2742|M.thermoresistible__bu ILDAAATALANAEYDTAISHLINAPAKIANAFINGYASDLENADDYFPGL
Mvan_1121|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_5178|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1270|M.smegmatis_MC2_155 --------------------------------------------------
TH_2742|M.thermoresistible__bu FSDRGTFNFFFSEVPADIANVLRLTRPESEESTASITGIPGSEPAVQLAL
Mvan_1121|M.vanbaalenii_PYR-1 ----------------ADEAAEREFEMTRRAERQKRWTLIGDTRAIYGED
Mflv_5178|M.gilvum_PYR-GCK ----------------EKLAAEREFERARLAERQHRWTMIGDSRAVYGEK
MSMEG_1270|M.smegmatis_MC2_155 ----------------VAERNRRHAVIARRADRQHQWVLDGDPRGIYGSE
TH_2742|M.thermoresistible__bu KSAPEQKVARDAQSKVEKDAPETPATQEPATEETAHQEPVTDETAPESSE
. ::. : * . ...
Mvan_1121|M.vanbaalenii_PYR-1 GAAAMRKITDDPEI----------DAEAAPAQDDSPFAQLATTSAELDAL
Mflv_5178|M.gilvum_PYR-GCK SSAPTADITG---------------TDSAPDT-DEPIATLAATKAELDAL
MSMEG_1270|M.smegmatis_MC2_155 GAEFMRYVER----------------------------------DNWDGL
TH_2742|M.thermoresistible__bu GTTGGELITSGASRNVRKGTSLKQWQDKAAERRQLRTEKVQQGLQNLQSN
.: : : :.
Mvan_1121|M.vanbaalenii_PYR-1 VR-------------EKPSAWPHALFASILVQRAAPLQSRLRDSELGFTA
Mflv_5178|M.gilvum_PYR-GCK IR-------------DRPQGWEQALFASILMQRAAPLASRLRDSELGYTE
MSMEG_1270|M.smegmatis_MC2_155 LRW-----------IQRPR-W-----------------------------
TH_2742|M.thermoresistible__bu VRKNLGFNRGGTDSQDTGAANNNAGTSKVXXXGGAPLQDRLRDSALGFTK
:* :
Mvan_1121|M.vanbaalenii_PYR-1 PTSTVALTPVHLASTVVTLIDEMIATAHQLEGFMAAPAFMAAFAGKDGEE
Mflv_5178|M.gilvum_PYR-GCK AGASGLMSAGELASTVVHMLDEMMRTIQQLDRFMMAPAFMAAFTAPDDVD
MSMEG_1270|M.smegmatis_MC2_155 --------------------------------------------------
TH_2742|M.thermoresistible__bu PTGERLITGWEVGGFIAELLDDFILLIDGVESFMRTPAFTNMFGAPGDEG
Mvan_1121|M.vanbaalenii_PYR-1 ---ADPEAIKHIAHRTMDYLERFMELSERCRALPVRSEHVDIVADCARML
Mflv_5178|M.gilvum_PYR-GCK EDGADPEAIAHIAHRVMDYHERLLELSERCRGISAPAYYDDILADCARML
MSMEG_1270|M.smegmatis_MC2_155 --------------------------------------------------
TH_2742|M.thermoresistible__bu --TADADGIVHTAHRLMDYHDRFLALSERCREAHAASRYIELLKDCARLT
Mvan_1121|M.vanbaalenii_PYR-1 DDPLASYREFIADFVDIIEALPRVLTHATGPVDMGSLGLYLKIDDKRHSR
Mflv_5178|M.gilvum_PYR-GCK DAPLRSYREFIDEFAAVIRALPRVLPHASGPVDLGSLGLYISIDNTLHSR
MSMEG_1270|M.smegmatis_MC2_155 --------------------------------------------------
TH_2742|M.thermoresistible__bu DAPLRGFQRFIDDFVARVEEMPVLLRYATGTVEVEPIVLHIDSDDALLRR
Mvan_1121|M.vanbaalenii_PYR-1 VLTRLDAITKS---
Mflv_5178|M.gilvum_PYR-GCK ILRRLDEITAPRRA
MSMEG_1270|M.smegmatis_MC2_155 --------------
TH_2742|M.thermoresistible__bu IVKQINKIPNN---