For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDSGAARVAVYLDFDNIVISRYDQVNGRNSFQRDKAKGPDDWAERVARATVDVGAIIDFASSFGTLVLTR AYADWSVEINAGYRGQLVQRAVDLVQLFPAAAYGKNGADIRLAVDAVEDMFRLPDLTHVVIVGGDSDYIA LAQRCKRLGRYVVGIGVAGSSSRMLAAACDEFVTYDALPGIPVMEPPPVADEKPAPKRRTSRAKAAEVAE TAEEPDSPDPQVAATGLLTRALRIGLEKEDVEWLHNSAVKAQMKRMDPSFSEKSLGFKSFSDFLRSRLDV VELDESSTTRLVRLREKP*
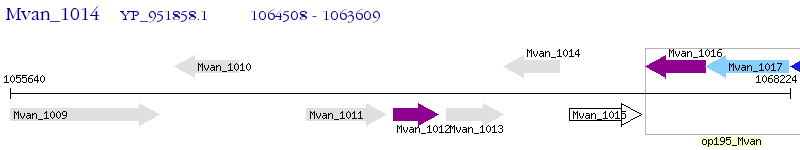
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1014 | - | - | 100% (299) | hypothetical protein Mvan_1014 |
| M. vanbaalenii PYR-1 | Mvan_5982 | - | 7e-05 | 28.12% (128) | hypothetical protein Mvan_5982 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4443 | - | 1e-146 | 87.00% (300) | hypothetical protein Mflv_4443 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2590 | - | 1e-127 | 79.32% (295) | hypothetical protein MMAR_2590 |
| M. avium 104 | MAV_4568 | - | 1e-130 | 81.25% (288) | hypothetical protein MAV_4568 |
| M. smegmatis MC2 155 | MSMEG_1210 | - | 1e-129 | 79.73% (296) | hypothetical protein MSMEG_1210 |
| M. thermoresistible (build 8) | TH_1870 | - | 1e-124 | 78.47% (288) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3170 | - | 1e-125 | 78.31% (295) | hypothetical protein MUL_3170 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1014|M.vanbaalenii_PYR-1 ---------------------------MTDSGAARVAVYLDFDNIVISRY
Mflv_4443|M.gilvum_PYR-GCK MGARTATFGPIRRHWGAAGYPSARLGAMTESGASRVAVYLDFDNIVLSRY
MAV_4568|M.avium_104 -----------------------------------MAVYLDFDNIVISRY
MSMEG_1210|M.smegmatis_MC2_155 ---------------------------MTEPAPTRVAVYLDFDNIVISRY
TH_1870|M.thermoresistible__bu -----------------------------------VAVYLDFDNIVISRY
MMAR_2590|M.marinum_M ---------------------------MTDSSTSRVAVYFDFDNIVISRY
MUL_3170|M.ulcerans_Agy99 ---------------------------MTDSSTSRVAVYFDFDNIVISRY
:***:******:***
Mvan_1014|M.vanbaalenii_PYR-1 DQVNGRNSFQRDKAKGPDDWAERVARATVDVGAIIDFASSFGTLVLTRAY
Mflv_4443|M.gilvum_PYR-GCK DQVNGRNSFQRDKAKGFDEVRDKLDRATVDVGAIIDFASSFGTLVLTRAY
MAV_4568|M.avium_104 DQIHGRNSFQRDKAKGLEQFTERLEQATVDVGAILDFASSFGTLVLTRAY
MSMEG_1210|M.smegmatis_MC2_155 DQVNGRNSFQKDKAKGLE--ADKLIKATVDVGAILDFASSFGTLVLTRAY
TH_1870|M.thermoresistible__bu DQLHGRNAFQKDRAAGFDQYPDRLAAATVDIGAIIDYASSFGTLVLTRAY
MMAR_2590|M.marinum_M EQVHGRNTFHRDKSKGLEQ--ERLQLATVDLGAIIDFASSFGTLVLTRAY
MUL_3170|M.ulcerans_Agy99 EQVHGRNTFHRDKSKGLEQ--ERLQLATVDLGAIIDFASSFGTLVLTRAY
:*::***:*::*:: * : ::: ****:***:*:*************
Mvan_1014|M.vanbaalenii_PYR-1 ADWSVEINAGYRGQLVQRAVDLVQLFPAAAYGKNGADIRLAVDAVEDMFR
Mflv_4443|M.gilvum_PYR-GCK ADWSAEINARYRGQLVGRAVDLVQLFPAAAYGKNGADIRLAVDAVEDMFR
MAV_4568|M.avium_104 ADWSAEINAGYRGQLVARAVDLVQLFPAAAYGKNGADIRLAVDAVEDMFR
MSMEG_1210|M.smegmatis_MC2_155 ADWSADVNAGYRQQLVGRAVDLVQLFPAAAYGKNGADIRLAVDAVEDMFR
TH_1870|M.thermoresistible__bu ADWSTEVNAAYRGQLVSRAVDLVQLFPTAAYGKNGADIRLAVDAVEDMFR
MMAR_2590|M.marinum_M ADWSADVNAGYHGQLVGRAVDLVQLFPAASYGKNGADIRLAVDAVEDMFR
MUL_3170|M.ulcerans_Agy99 ADWSADVNAGYHGQLVGRAVDLVQLFPAASYGKNGANIRLAVDAVEDMFR
****.::** *: *** **********:*:******:*************
Mvan_1014|M.vanbaalenii_PYR-1 LPDLTHVVIVGGDSDYIALAQRCKRLGRYVVGIGVAGSSSRMLAAACDEF
Mflv_4443|M.gilvum_PYR-GCK LPDLTHVVIVAGDSDYIALAQRCKRLGRYVVGIGVAGSSSRMLAAACDEF
MAV_4568|M.avium_104 LPDLTHVVIVAGDSDYIPLAQRCKRLGRYVVGIGVAGASSRALAAACDEF
MSMEG_1210|M.smegmatis_MC2_155 LPDLTHVVIVAGDSDYIPLAQRCKRLGRYVVGIGVAGASSRALAAACDEF
TH_1870|M.thermoresistible__bu LPDLTHVVIVAGDSDYIPLAQRCRRLGRYVVGIGVAGSISRSLTAACDEY
MMAR_2590|M.marinum_M LPDLTHVVIVGGDSDYIALAQRCKRLGRYVVGIGVAGASSGSLAAACDEF
MUL_3170|M.ulcerans_Agy99 LPDLTHVVIVGGDSDYIALAQRCKRLGRYVVGIGVAGASSGSLAAACDEF
**********.******.*****:*************: * *:*****:
Mvan_1014|M.vanbaalenii_PYR-1 VTYDALPGIPVMEPPPVADEKPAPKRRTSRAKAA---EVAETAEEPDSPD
Mflv_4443|M.gilvum_PYR-GCK VTYDTLPGVPVFEPEPVADEKPAPKRRSTRAKATSEEAATETPEEPEQPD
MAV_4568|M.avium_104 VIYDALPGVTALDRTPAPAETVPAKPRTRRGKAA----------QPEEPD
MSMEG_1210|M.smegmatis_MC2_155 IIYDALPGVPVFEPEPQTTPARRTR-RTKKDENE----------EPEPPD
TH_1870|M.thermoresistible__bu VTYDALPGVP-VESTPAPKRRPRRKAVASDADGD-----------DDAHD
MMAR_2590|M.marinum_M VTYDALPGIPTLPPPAAKKRS----RRSTAADRD----------EPTAPD
MUL_3170|M.ulcerans_Agy99 VTYDALPGIPTLPPPAAKKRS----RRSTAGDRD----------EPTPPD
: **:***:. . . : . *
Mvan_1014|M.vanbaalenii_PYR-1 PQVAATGLLTRALRIGLEKEDVEWLHNSAVKAQMKRMDPSFSEKSLGFKS
Mflv_4443|M.gilvum_PYR-GCK PQTSATGLLTRALRIGLEKDDADWLHNSAVKAQMKRMDPSFSEKSLGFKS
MAV_4568|M.avium_104 PQAAATALLTRALQIGLEKDDVDWLHNSAVKAQMKRMDPSFSEKSLGFRS
MSMEG_1210|M.smegmatis_MC2_155 PQATATALLLRALQIGLEKDDVEWLHNSAVKAQMKRMDPSFSEKSLGFRS
TH_1870|M.thermoresistible__bu PQEAATNLLLRALRIGLERDDVEWLHNSAVKAQMKRMDPSFSEKSLGYRS
MMAR_2590|M.marinum_M AQTAATGLLERALRIGQEKDDAEWLHNSAVKAQMKRMDPSFSEKSLGFKS
MUL_3170|M.ulcerans_Agy99 AQTAATGLLKRALRIGQEKDDAEWLHNSAVKAQMKRMDPSFSEKSLGFKS
.* :** ** ***:** *::*.:************************::*
Mvan_1014|M.vanbaalenii_PYR-1 FSDFLRSRLDVVELDESSTTRLVRLREKP--
Mflv_4443|M.gilvum_PYR-GCK FSDFLRSRSDVVELDESSTIRMVRLRENA--
MAV_4568|M.avium_104 FSDFLRSRSDVVELDETSTTRMVRLRPHE--
MSMEG_1210|M.smegmatis_MC2_155 FSDFLRSRSDYVELDESSTIRMVRLRPDR--
TH_1870|M.thermoresistible__bu FSDFLRSRGDLVELDESSTTRLVRLRTDD--
MMAR_2590|M.marinum_M FSDFLRSRSDLVDLDESSTTRLVRLHTEADS
MUL_3170|M.ulcerans_Agy99 FSDFLRSRSDLVDPDESSTTRLVRLHTEADS
******** * *: **:** *:***: .