For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SREVTSVEELREIVGHPNAVVANKVRDRLSPVEQHWLSHSPLGFVATMDAHGRLDISPKGDPAGFVQIID DRTIAIPERPGNRRVDGYLNVLERPRVGTLFVIPGRGDTLRINGSARIMADADYFDAMTVQGKRPILALQ VAVEEVFFHCAKAFLRSDAWQPETWNPTAVPSAAQLAKAIYADADLDEIRKSLDEENLRKVLY*
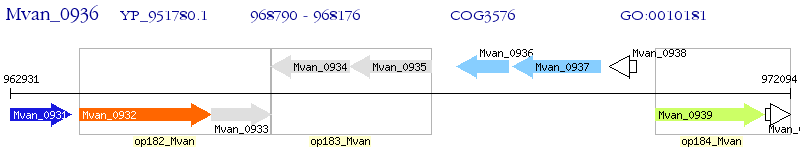
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0936 | - | - | 100% (204) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
| M. vanbaalenii PYR-1 | Mvan_4731 | - | 6e-06 | 26.16% (172) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0034 | - | 6e-96 | 79.90% (204) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2382 | - | 1e-32 | 39.10% (156) | hypothetical protein MAB_2382 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1061 | - | 1e-84 | 72.06% (204) | phosphohydrolase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0936|M.vanbaalenii_PYR-1 ------------------MSREVTSVEELREIVGHPNAVVANKVRDRLSP
Mflv_0034|M.gilvum_PYR-GCK ------------------MSRHVTTVEELRAIVGHPHAVVAGKVRDRLLP
MSMEG_1061|M.smegmatis_MC2_155 ------------------MRQEVTSVDELRAIVGHPNKYVANKVTARLSE
MAB_2382|M.abscessus_ATCC_1997 MSATPTDTSPPHPPETAADRYSPITMDRVREVIGFPARFIADKKDTELRE
:::.:* ::*.* :*.* .*
Mvan_0936|M.vanbaalenii_PYR-1 VEQHWLSHSPLGFVATMDAHGRLDISPKGDPAGFVQIIDDRTIAIPERPG
Mflv_0034|M.gilvum_PYR-GCK AMRDWLAHSPLGFVATTDADGRVDVSPKGDPAGFVHIIDDHTIAIPERPG
MSMEG_1061|M.smegmatis_MC2_155 VQQDWLAASPLCFVATTDAEGRVDVSPKGDPPGFVRVIDDTTIAIPERPG
MAB_2382|M.abscessus_ATCC_1997 FAARFIAHSPFFCMATVGADGLLDTSPKGDPPGSVRILDARTLAIPDRPG
::: **: :** .*.* :* ******.* *:::* *:***:***
Mvan_0936|M.vanbaalenii_PYR-1 NRRVDGYLNVLERPRVGTLFVIPGRGDTLRINGSARIMADADYFDAMTVQ
Mflv_0034|M.gilvum_PYR-GCK NKRVDGYLNVLERPRVGTLFVIPGRGDTLRINGSARVMADADYFDAMSVE
MSMEG_1061|M.smegmatis_MC2_155 NKRVDGYLNVLQQPHVGTVFLIPGRGDTLRINGTATILSDADYFDDMVVD
MAB_2382|M.abscessus_ATCC_1997 NKLADSFENITRNPAVGLVFFVPGLREVIRVNGDAFVTDDPELLAMLSAD
*: .*.: *: ..* ** :*.:** :.:*:** * : *.: : : .:
Mvan_0936|M.vanbaalenii_PYR-1 GKRPILALQVAVEEVFFHCAKAFLRSDAWQPETWNPTAVPSAA----QLA
Mflv_0034|M.gilvum_PYR-GCK GRRPILALEIAVEEVFFHCAKAFLRSDAWDPQSWDPAAVPSAA----HLA
MSMEG_1061|M.smegmatis_MC2_155 GKRPILALEISIEEVFFHCAKAFLRSDTWKPETWNPTAVPSVA----QLA
MAB_2382|M.abscessus_ATCC_1997 GKPAVLATIVRIREVFGQCGKAVIRAKLWEGDQRGLAEAVTLGGDFYTLT
*: .:** : :.*** :*.**.:*:. *. : . : . : . *:
Mvan_0936|M.vanbaalenii_PYR-1 KAIYADADLDEIRKSLD------EENLRKVLY
Mflv_0034|M.gilvum_PYR-GCK KQMYADADLDDMRRALD------EDMMRQALY
MSMEG_1061|M.smegmatis_MC2_155 KAFKPDMSDKELDEYYA------EDNLRKILY
MAB_2382|M.abscessus_ATCC_1997 IAENAAKMADKLGQLAGGIHDLVDDSYANELF
. ..: . :: : *: