For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AAPIGQVTRGTTGHNRLRRSDRWLVHSSRVRTALQTTSDPLVVDLGYGAKPVTTLELAIRLRQVRADVRV VGLEIDPERVRAGQAAATPGVEFALGGFELAGRRPVLVRAFNVLRQYPVEAVHDAWTAMQHRVSAGGLIV DGTCDELGRLCCWILLDAAAPVSLTLACDPFAIERPSDLAVRLPKILIHHNVEGQPIHALLRAADRAWAS VAGHGVFGPRVRWRAMLELLVGEGFPVAAPRRSLRDGILTVPWAAVAPTAR*
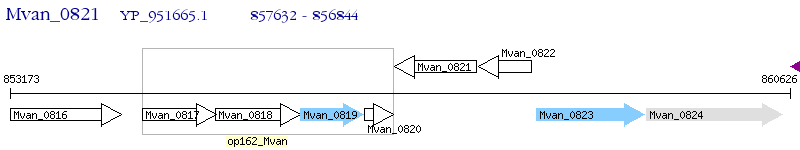
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0821 | - | - | 100% (262) | hypothetical protein Mvan_0821 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0092 | - | 1e-128 | 84.03% (263) | hypothetical protein Mflv_0092 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4075 | - | 3e-98 | 67.83% (258) | hypothetical protein MAB_4075 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0925 | - | 1e-116 | 78.82% (255) | hypothetical protein MSMEG_0925 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0821|M.vanbaalenii_PYR-1 ---MAAPIGQVTRGTTGHNRLRRSDRWLVHSSRVRTALQTTSDPLVVDLG
Mflv_0092|M.gilvum_PYR-GCK ----MAPLGQVTRGTTGHNRLRRSDRWLVHSSRVRTALQAAADPLVVDLG
MSMEG_0925|M.smegmatis_MC2_155 -------MGAVTRGTTGYNRLRRSDRWLVHEPRVRTALRSASDPLVIDLG
MAB_4075|M.abscessus_ATCC_1997 MAPPARPRGTITRGTTGINRLRRSDRWLVHHPGVQAALASAADPLVVDLG
* :****** ************ . *::** :::****:***
Mvan_0821|M.vanbaalenii_PYR-1 YGAKPVTTLELAIRLRQVRADVRVVGLEIDPERVRAGQAAA----TPGVE
Mflv_0092|M.gilvum_PYR-GCK YGAKPVTTLELAIRLREVRRDVRVVGLEIEPERVRAGQSAVRHGAHPGVE
MSMEG_0925|M.smegmatis_MC2_155 YGALPITTLELASRLQIVRADLRVVGLEIHPERVATARALA----APTVQ
MAB_4075|M.abscessus_ATCC_1997 YGAMPNTTLELAARLRTVRSDIRVVGLEIDPQRVVPA--------RDGVH
*** * ****** **: ** *:*******.*:** .. *.
Mvan_0821|M.vanbaalenii_PYR-1 FALGGFELAGRRPVLVRAFNVLRQYPVEAVHDAWTAMQHRVSAGGLIVDG
Mflv_0092|M.gilvum_PYR-GCK FALGGFELADHRPILVRAFNVLRQYPVEDVEGAWAAMRDRLATGGLIVDG
MSMEG_0925|M.smegmatis_MC2_155 FALGGFELAGLRPVLVRAFNVLRQYPVEAVPDAWATMRRQLSPGGLIVDG
MAB_4075|M.abscessus_ATCC_1997 FGLGGFELAGLTPVLVRAFNVLRQYPESEVAQAWATMTDRLAPGGLIIEG
*.*******. *:************ . * **::* :::.****::*
Mvan_0821|M.vanbaalenii_PYR-1 TCDELGRLCCWILLDAAAPVSLTLACDPFAIERPSDLAVRLPKILIHHNV
Mflv_0092|M.gilvum_PYR-GCK TCDELGRLCCWVLLDATRPVSLTLACDPFAIERPSQLAERLPKVLIHHNV
MSMEG_0925|M.smegmatis_MC2_155 TCDELGRRCCWVLLDADGPVSLTLACDPFAIERPSDLAERLPKVLIHHNV
MAB_4075|M.abscessus_ATCC_1997 TCDEIGRRCAWLLLDASGPLTLTLACDPFDIDKPSDLAERLPKALIHHNV
****:** *.*:**** *::******** *::**:** **** ******
Mvan_0821|M.vanbaalenii_PYR-1 EGQPIHALLRAADRAWASVAGHGVFGPRVRWRAMLELLVGEGFPVAAPRR
Mflv_0092|M.gilvum_PYR-GCK EGQPIHTLLRAADRAWASVAGHGVFGPRIRWRAMLDLLVAEGFPIEVPRR
MSMEG_0925|M.smegmatis_MC2_155 AGQPIHTLLTAADRAWASVAGHGVFGPRVRWRAMLELLRDEGFGVQPPRR
MAB_4075|M.abscessus_ATCC_1997 PSQPIHDLLAAADKYWAAAAPHGVFGPRVRWRMMLKALRDNGFPVEQQRR
.**** ** ***: **:.* *******:*** **. * :** : **
Mvan_0821|M.vanbaalenii_PYR-1 SLRDGILTVPWAAVAPTAR---
Mflv_0092|M.gilvum_PYR-GCK SLRDGVLTVPWDAVAPVS----
MSMEG_0925|M.smegmatis_MC2_155 RMRDGVLTVSWQTIAPLKN---
MAB_4075|M.abscessus_ATCC_1997 QVRDCILTTPWSAVAPVTGPSR
:** :**..* ::**