For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDAPNRALVTEAAAELLRSLQDRHGALMFHQSGGCCDGSSPMCYPDGEFLVGDRDVLLAVLDVGDGVPVW ISGPQFDAWKHTQLVIDVVPGRGGGFSLEAPEGKRFLSRGRAFTEEENRALDAVPPVTGAAYERGERPVS NGTHVVAEAQAVDACAIPLPE
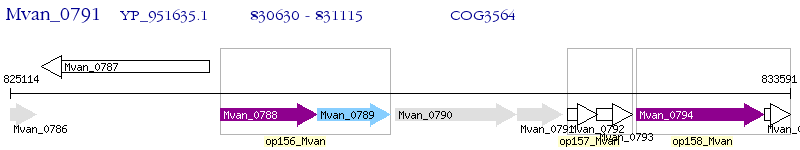
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0791 | - | - | 100% (161) | hypothetical protein Mvan_0791 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0468 | - | 1e-58 | 72.03% (143) | hypothetical protein Mb0468 |
| M. gilvum PYR-GCK | Mflv_0122 | - | 2e-75 | 84.52% (155) | hypothetical protein Mflv_0122 |
| M. tuberculosis H37Rv | Rv0459 | - | 1e-58 | 72.03% (143) | hypothetical protein Rv0459 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0973 | - | 4e-40 | 62.70% (126) | hypothetical protein MAB_0973 |
| M. marinum M | MMAR_0781 | - | 5e-60 | 69.33% (163) | hypothetical protein MMAR_0781 |
| M. avium 104 | MAV_4690 | - | 5e-55 | 70.00% (150) | hypothetical protein MAV_4690 |
| M. smegmatis MC2 155 | MSMEG_0901 | - | 1e-69 | 76.58% (158) | hypothetical protein MSMEG_0901 |
| M. thermoresistible (build 8) | TH_0971 | - | 1e-48 | 71.65% (127) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1417 | - | 1e-59 | 68.71% (163) | hypothetical protein MUL_1417 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0791|M.vanbaalenii_PYR-1 -------------------------MDAPNRALVTEAAAELLRSLQDRHG
Mflv_0122|M.gilvum_PYR-GCK -------------------------MAVPHRALITAAAAELLLTLQERHG
MSMEG_0901|M.smegmatis_MC2_155 -------------------------MSTPSRALITAAAADLLARLQKTHG
MMAR_0781|M.marinum_M ----------------------MPEPSAPSGVVITAGAAELLARLQDRHG
MUL_1417|M.ulcerans_Agy99 ----------------------MPEPSAPSGVVITAGAAELLARLQDRHG
MAV_4690|M.avium_104 -----------------------MEPRPPSGVVITAAAAEVLARLQRQHG
Mb0468|M.bovis_AF2122/97 -------------------------MNAPAGVLITAEAAALLAGLQDRHG
Rv0459|M.tuberculosis_H37Rv -------------------------MNAPAGVLITAEAAALLAGLQDRHG
MAB_0973|M.abscessus_ATCC_1997 -----------MPRASSDEGSPEESGILINRVELTPAAAELLASLVAKHG
TH_0971|M.thermoresistible__bu MGPDGDFGVETFAQRPRARRNREEKEVPPPRVDITGAAAELVRRLRARHG
. :* ** :: * **
Mvan_0791|M.vanbaalenii_PYR-1 ALMFHQSGGCCDGSSPMCYPDGEFLVGDRDVLLAVLDVG-----DGVPVW
Mflv_0122|M.gilvum_PYR-GCK PLMFHQSGGCCDGSSPMCYPAGDFIVGDRDVLLAVLDVG-----TGVPVW
MSMEG_0901|M.smegmatis_MC2_155 ALMFHQSGGCCDGSSPMCYPEGDFIVGDRDVLLGVLDVG-----DGVPVW
MMAR_0781|M.marinum_M PVMFHQSGGCCDGSSPMCYPRGDFLVGDRDVLLGTLDIGKQTGAEGVAVW
MUL_1417|M.ulcerans_Agy99 PVMFHQSGGCCDGSSPMCYPRGDFLVGDRDVLLGTLDIGKQTGAEGVAVW
MAV_4690|M.avium_104 PVMFHQSGGCCDGSSPMCYPVGDFLVGDRDVLLGVLDVG----ADGVPVW
Mb0468|M.bovis_AF2122/97 PVMFHQSGGCCDGSAPMCYPRADFLVGDRDILLGVLDVG----EDGVPVW
Rv0459|M.tuberculosis_H37Rv PVMFHQSGGCCDGSAPMCYPRADFLVGDRDILLGVLDVG----EDGVPVW
MAB_0973|M.abscessus_ATCC_1997 PVMFHQSGGCCDGSAPMCYPRGEFRVGASDVLLGEVSRD-------TPFW
TH_0971|M.thermoresistible__bu PLMFHQSGGCCDGSLPMCFPEGEFLVGDRDVLLGVVEQ--------TPVW
.:************ ***:* .:* ** *:**. :. ...*
Mvan_0791|M.vanbaalenii_PYR-1 ISGPQFDAW---KHTQLVIDVVPGRGGGFSLEAPEGKRFLSRGRAFTEEE
Mflv_0122|M.gilvum_PYR-GCK ISGPQFDTW---KHTQLVIDVVPGRGGGFSLESPEGKRFLSRGRAFTEDE
MSMEG_0901|M.smegmatis_MC2_155 ISGPQFEAW---KHTQLVIDVVPGRGGGFSLESPEGMRFLSRGRAFTDAE
MMAR_0781|M.marinum_M ISGPQYQAW---KHTQLVIDVVPGRGGGFSLEAPEGMRFLSRGRVFTGEE
MUL_1417|M.ulcerans_Agy99 ISGPQYQAW---KHTQLVIDVVPGRGGGFSLEAPEGMRFLSRGRVFTGEE
MAV_4690|M.avium_104 ISGPQYAAHYRDKHTQLVIDVVPGRGGGFSLEAPDGVRFLSRGRVFSAAE
Mb0468|M.bovis_AF2122/97 ISGPQYQAW---KHTQLIIDVVPGRGGGFSLEAPEGVRFLSRGRVFSDAE
Rv0459|M.tuberculosis_H37Rv ISGPQYQAW---KHTQLIIDVVPGRGGGFSLEAPEGVRFLSRGRVFSDAE
MAB_0973|M.abscessus_ATCC_1997 MSADQFEYW---SHTHLTVDVVPGRGSGFSLEAPEGVRFLIRSRLFTDDE
TH_0971|M.thermoresistible__bu ISGPQFDAW---RHTQLIIDVVPGRGAGFSLEAPEGVRFLTRGRAFDERE
:*. *: **:* :*******.*****:*:* *** *.* * *
Mvan_0791|M.vanbaalenii_PYR-1 NRALDAVPPVTGAAYERGERPVSNGTHV--VAEAQAVDACAIPLPE
Mflv_0122|M.gilvum_PYR-GCK NRALDDAPPVTGAAYERGERPSPTGSHL--VAD--AVDACAIPQK-
MSMEG_0901|M.smegmatis_MC2_155 NEELAAQPPITGAQYAEGARPVQTGTSV--VAE--AEDACPVPGR-
MMAR_0781|M.marinum_M QALLEETPVITGAAYERGERPAVPGEVV----ADNAPGACPASRP-
MUL_1417|M.ulcerans_Agy99 QALLEETPVITGAVYERGERPAVPGEVV----ADNAPGACPASRP-
MAV_4690|M.avium_104 QAAVAAAPIITGADYQHGERPAARGAVV----ADAACRTCP-----
Mb0468|M.bovis_AF2122/97 KAMREAAPVITGAAYECGERPLVRGLVVDLDDPDATPGVCRASRR-
Rv0459|M.tuberculosis_H37Rv KAMREAAPVITGAAYECGERPLVRGLVVDLDDPDATPGVCRASRR-
MAB_0973|M.abscessus_ATCC_1997 VLALANQPVRTGADG-------------------------------
TH_0971|M.thermoresistible__bu IRLLADRRVLTGADCAGAGRTPGVER--------------------
***