For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTETNPTATVVAYEPSLGRPARAGSWDELAKGTFRVATEHLVAGDWDAAAALVEIAVVEAEELRDVYERW PVSTRGWIESRGVTADLVDAATARLTVMIGERAMAGIEAEWPQFVAAVDRAALACRAQLPDAAAAVETAR AVWQGVHDRAVDRVSGLIDIAVSTVGEHSLGELWDALMADWYDVHEQRYALSNQPWEASAHQLMVAIVDG FHAHLTGTGRQGDMEIIDEPTRIGFRFAPCGSGGRSLDARITDGTPRAGAPFGFAVTTAPHDWAWNTVGI CSYCVHCCQLNEVMPIDRLGYPTRVIDAPTWNADNPVTECTWWVYRDPADVPDHVYERVGRSPDRRPTRR GDVK
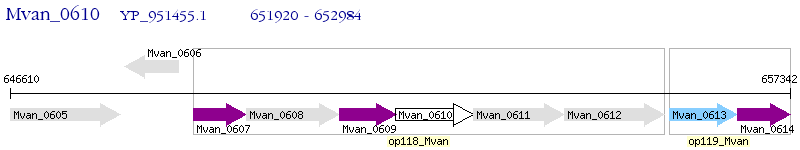
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0610 | - | - | 100% (354) | hypothetical protein Mvan_0610 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3826 | - | 1e-155 | 71.88% (352) | hypothetical protein MSMEG_3826 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0610|M.vanbaalenii_PYR-1 MTETNPTATVVAYEPSLGRPARAGSWDELAKGTFRVATEHLVAGDWDAAA
MSMEG_3826|M.smegmatis_MC2_155 MTRTPVTRNPVAYEPALGRNARTGDWQELARGTFRTAIERVEAQQWEAAA
**.* * . *****:*** **:*.*:***:****.* *:: * :*:***
Mvan_0610|M.vanbaalenii_PYR-1 ALVEIAVVEAEELRDVYERWPVSTRGWIESRGVTADLVDAATARLTVMIG
MSMEG_3826|M.smegmatis_MC2_155 QLVEVAVLEAEELNDVYQRWPAATMQWIRDHDVAQADLDRALARLTALIG
***:**:*****.***:***.:* **..:.*: :* * ****.:**
Mvan_0610|M.vanbaalenii_PYR-1 ERAMAGIEAEWPQFVAAVDRAALACRAQLPDAAAAVETARAVWQGVHDRA
MSMEG_3826|M.smegmatis_MC2_155 DQAMAGINAEWPTFTDAVAVAARACRDQDPAAAGLIETARQAWQQIHDRA
::*****:**** *. ** ** *** * * **. :**** .** :****
Mvan_0610|M.vanbaalenii_PYR-1 VDRVSGLIDIAVSTVGEHSLGELWDALMADWYDVHEQRYALSNQPWEASA
MSMEG_3826|M.smegmatis_MC2_155 VDRVAGIVDIAVTLVGEPALGELWDFLMADWYEIHERRYALDNQPWSESA
****:*::****: *** :****** ******::**:****.****. **
Mvan_0610|M.vanbaalenii_PYR-1 HQLMVAIVDGFHAHLTGTGRQGDMEIIDEPTRIGFRFAPCGSGGRSLDAR
MSMEG_3826|M.smegmatis_MC2_155 HQLMIAIVDGFHAHLAGTGRQGDIELIEEPGRTGFRFAPCGSGGRSLDAR
****:**********:*******:*:*:** * *****************
Mvan_0610|M.vanbaalenii_PYR-1 ITDGTPRAGAPFGFAVTTAPHDWAWNTVGICSYCVHCCQLNEVMPIDRLG
MSMEG_3826|M.smegmatis_MC2_155 ITDGVPRSGAPFGFAVTTEPHDWAWNTVGICSYCVHCCQLNEVMPIDRLG
****.**:********** *******************************
Mvan_0610|M.vanbaalenii_PYR-1 YPTRVIDAPTWNADNPVTECTWWVYRDPADVPDHVYERVGRSPDRRP--T
MSMEG_3826|M.smegmatis_MC2_155 YPTRVIDPPTWTPDSPTTSCTWWVYHDLADIPDHVYHRVGRDPARRPSPT
*******.***..*.*.*.******:* **:*****.****.* *** *
Mvan_0610|M.vanbaalenii_PYR-1 RRGDVK
MSMEG_3826|M.smegmatis_MC2_155 RRSADG
**.