For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVSVTDLGIDPAAGFTSVDEANATAFERLDAADPWVVDVAPARDVLPGYRDNLVLTSGAPMAWPDYTGGQ REALIGGALFEGLADSRDDAIAGFDSGRIEVGACQDHAAVGSLAGIYTASMPVFVVENRTHGNRGFCNFY EGKEPRRLNYGCYDEGVHERLLHVNTVLAPVVGEAIRRRGGIALKPLIARALRMGDEVHSRNAAASILFN REIMPAILDMCDDRVPGVRETMLHLAENDYFFLRLSMAAAKASADAMVVPGSTLISAMAFSCRGFAVKLA GLGDTWFEGPPPIHQGKLFAGHTEDEITWMGGESPITETVGLGGFAQACALSLQEYQGGSPEVMIERNLE MYSITHGENSTYRIPQFGFRGTPTGIDARKVLQTGVLPVMDVGLAGRDGGQIGAGVIRAPRECFADALAE HDRRFGAR
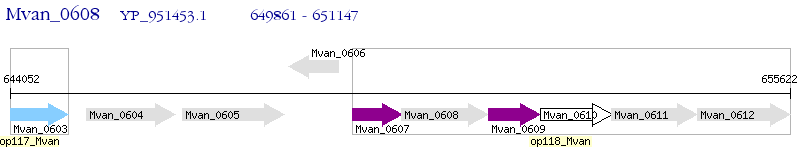
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0608 | - | - | 100% (428) | hypothetical protein Mvan_0608 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3824 | - | 0.0 | 88.51% (409) | hypothetical protein MSMEG_3824 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0608|M.vanbaalenii_PYR-1 MVSVTDLGIDPAAGFTSVDEANATAFERLDAADPWVVDVAPARDVLPGYR
MSMEG_3824|M.smegmatis_MC2_155 MTTEIHTPTAATDGID-VEAANATAFARLDEADPWVIDVRPAREVLPGYR
*.: . .: *: *: ****** *** *****:** ***:******
Mvan_0608|M.vanbaalenii_PYR-1 DNLVLTSGAPMAWPDYTGGQREALIGGALFEGLADSRDDAIAGFDSGRIE
MSMEG_3824|M.smegmatis_MC2_155 DNVVLTSGAPMPWSAYTGGQREALIGGALFEGLAATREEAIAKFDDGTIE
**:********.*. ******************* :*::*** **.* **
Mvan_0608|M.vanbaalenii_PYR-1 VGACQDHAAVGSLAGIYTASMPVFVVENRTHGNRGFCNFYEGKEPRRLNY
MSMEG_3824|M.smegmatis_MC2_155 VGACHDYGAVGSLAGIYTASMPVFVVENRAHGNLGFCNFYEGKEPRRLNY
****:*:.*********************:*** ****************
Mvan_0608|M.vanbaalenii_PYR-1 GCYDEGVHERLLHVNTVLAPVVGEAIRRRGGIALKPLIARALRMGDEVHS
MSMEG_3824|M.smegmatis_MC2_155 GCYDAGVHERLEHVNTVLAPVVGEAIRRQGGIALKPLIARALRMGDEVHS
**** ****** ****************:*********************
Mvan_0608|M.vanbaalenii_PYR-1 RNAAASILFNREIMPAILDMCDDRVPGVRETMLHLAENDYFFLRLSMAAA
MSMEG_3824|M.smegmatis_MC2_155 RNAAASILFNREILPAVLDMVGEGVAGVRETMLHLAENDYFFLRLSMAAA
*************:**:*** .: *.************************
Mvan_0608|M.vanbaalenii_PYR-1 KASADAMVVPGSTLISAMAFSCRGFAVKLAGLGDTWFEGPPPIHQGKLFA
MSMEG_3824|M.smegmatis_MC2_155 KASADAMVTPGSTLVSAMAFSCRGFAIKLAGLGDQWFEGPPPIHQGKLFA
********.*****:***********:******* ***************
Mvan_0608|M.vanbaalenii_PYR-1 GHTEDEITWMGGESPITETVGLGGFAQACALSLQEYQGGSPEVMIERNLE
MSMEG_3824|M.smegmatis_MC2_155 GHSEDEITWMGGESPITETIGLGGFAQACALSLQEYQGGLPEVMIERNRE
**:****************:******************* ******** *
Mvan_0608|M.vanbaalenii_PYR-1 MYSITHGENSTYRIPQFGFRGTPTGIDARKVLQTGVLPVMDVGLAGRDGG
MSMEG_3824|M.smegmatis_MC2_155 MYAITHGENSTYRIPLFAFRGTPTGIDARKVLDTGVLPVMDVGLAGRDGG
**:************ *.**************:*****************
Mvan_0608|M.vanbaalenii_PYR-1 QIGAGVIRAPRECFADALAEHDRRFGAR
MSMEG_3824|M.smegmatis_MC2_155 QIGAGVIRAPRECFADAMAEHTRRFGRP
*****************:*** ****