For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSTSLQPKRRADDVTALRVMTEEMRPLLVGDGYQMFDVRAPKSSGPPPHQHPWDESYVVLEGELWVSRGG EEATLQVGEAIRVPAGVVHAYRVLSDGARWITTSSCPGGALDFFSDVDETSNGPGDVKALIEAAKRNSVR FADPALG
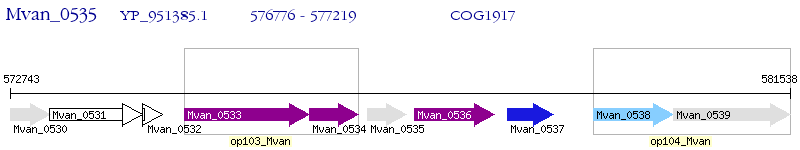
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0535 | - | - | 100% (147) | cupin 2 domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_0760 | - | 7e-05 | 31.58% (76) | cupin 2 domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0630 | - | 7e-83 | 99.32% (147) | cupin 2 domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0758 | - | 2e-05 | 35.71% (70) | hypothetical protein MMAR_0758 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6861 | - | 1e-05 | 33.33% (72) | cupin domain-containing protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1392 | - | 2e-05 | 35.71% (70) | hypothetical protein MUL_1392 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0758|M.marinum_M ------MSLVMPAYPPYPPARYTGEKGEVSARLKRRDEPPNLELGGTK--
MUL_1392|M.ulcerans_Agy99 MGHTGVMSLVMPAYPPYPPARYTGEKGEVSARLKRRDEPPNLELGGTK--
Mvan_0535|M.vanbaalenii_PYR-1 ------------MSTSLQPKR----RADDVTALRVMTEEMRPLLVGDG--
Mflv_0630|M.gilvum_PYR-GCK ------------MSTSLQPKR----RADDVTALRVMTEEMRPLLVGDG--
MSMEG_6861|M.smegmatis_MC2_155 ------------MTTDDKPAEVVLGQAGASPAYWYRAVLWNVLMSADQTL
. * . :. . . : .
MMAR_0758|M.marinum_M --YHYLATQKTTDGDFGLYRVDIAPAGGGPGPHFHKTMSESFFVLSGSMR
MUL_1392|M.ulcerans_Agy99 --YHYLATQKTTDGDFGLYRVDIAPAGGGPGPHFHKTMSESFFVLSGSMR
Mvan_0535|M.vanbaalenii_PYR-1 --YQMFDVR--------------APKSSGPPPHQHP-WDESYVVLEGELW
Mflv_0630|M.gilvum_PYR-GCK --YQMFDVR--------------APQSSGPPPHQHP-WDESYVVLEGELW
MSMEG_6861|M.smegmatis_MC2_155 GEFTLLEQ--------------VIPAGAGPPAHVHERQAEGFYVISGELE
: : * ..** .* * *.: *:.*.:
MMAR_0758|M.marinum_M MHDGRDWVDASAG--DYLYVPPGGVHGFRNESDQPASILILFAPGAP-RE
MUL_1392|M.ulcerans_Agy99 MHDGRDWVDASAG--DYLYVPPGGVHGFRNESDQPASILILFAPGAP-RE
Mvan_0535|M.vanbaalenii_PYR-1 VSRGGEEATLQVG--EAIRVPAGVVHAYRVLSDGARWITTSSCPGG--AL
Mflv_0630|M.gilvum_PYR-GCK VSRGGEEATLQVG--EAIRVPAGVVHAYRVLSDGARWITTSSCPGG--AL
MSMEG_6861|M.smegmatis_MC2_155 LVVGAADEVMRAGPGAAVWIPKSTRHAFRVVSDEDARVLNFYAPGGFDDH
: * .* : :* . *.:* ** : .**.
MMAR_0758|M.marinum_M DYFEGFPGLAD--------MTDDQRREFFVAHDN--FWVQ----------
MUL_1392|M.ulcerans_Agy99 DYFEGFPGLAD--------MTDDQRREFFVAHDN--FWVQ----------
Mvan_0535|M.vanbaalenii_PYR-1 DFFSDVDETSN--------GPGDVKALIEAAKRNSVRFADPALG------
Mflv_0630|M.gilvum_PYR-GCK DFFSDVDETSN--------GPGDVKALIEAAKRNSVRFADPALG------
MSMEG_6861|M.smegmatis_MC2_155 LPFYGVKATAQTLPPAELGAPVLDRERMATSQESRNAYLQRLADIAEGTW
* .. :: . : : .:: . : :
MMAR_0758|M.marinum_M ----------
MUL_1392|M.ulcerans_Agy99 ----------
Mvan_0535|M.vanbaalenii_PYR-1 ----------
Mflv_0630|M.gilvum_PYR-GCK ----------
MSMEG_6861|M.smegmatis_MC2_155 DFSIADPSAH