For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNDRDFVEKRFSRPGEYRAAVIYTFVVVVLAAIAFAVYAFGDRSSVFSAALVPVFLFAGGVGALLRTYRQ WKGGGGWTAWQGAGWFLLLLMLLTLAVPGGAAFSN*
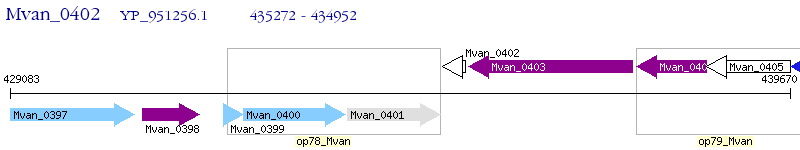
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0402 | - | - | 100% (106) | hypothetical protein Mvan_0402 |
| M. vanbaalenii PYR-1 | Mvan_1654 | - | 4e-05 | 40.00% (45) | hypothetical protein Mvan_1654 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3317c | - | 8e-12 | 49.15% (59) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_0340 | - | 1e-48 | 81.90% (105) | hypothetical protein Mflv_0340 |
| M. tuberculosis H37Rv | Rv3289c | - | 8e-12 | 49.15% (59) | transmembrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2521c | - | 5e-12 | 37.23% (94) | hypothetical protein MAB_2521c |
| M. marinum M | MMAR_1244 | - | 2e-12 | 37.37% (99) | hypothetical protein MMAR_1244 |
| M. avium 104 | MAV_4261 | - | 8e-11 | 53.33% (45) | hypothetical protein MAV_4261 |
| M. smegmatis MC2 155 | MSMEG_0601 | - | 6e-22 | 48.57% (105) | hypothetical protein MSMEG_0601 |
| M. thermoresistible (build 8) | TH_0798 | - | 7e-24 | 51.04% (96) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2643 | - | 5e-12 | 38.89% (90) | hypothetical protein MUL_2643 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3317c|M.bovis_AF2122/97 MHEVGGPSRGDRLGRDDSEVHSAIRFAVVAAVVGVGFLIMGALLVSTCSG
Rv3289c|M.tuberculosis_H37Rv MHEVGGPSRGDRLGRDDSEVHSAIRFAVVAAVVGVGFLIMGALLVSTCSG
MMAR_1244|M.marinum_M MQRVHERSDGDRLGQDDEEVRAAIRFAALAASAGVGFLIMAALWVSTCGG
MUL_2643|M.ulcerans_Agy99 MQRVHERSDGDRLGQDDEQVRAAIRFAALAASAGVGFLIMAALWVSTCGG
MAV_4261|M.avium_104 -------------------MHAAIRFAVLCAAGAVGFLLIAAVWVSTCPT
MAB_2521c|M.abscessus_ATCC_199 -MNAPMYSDAERKDRDKRTHIHAVRFSAATVALAVVTFVTAIVWMGQCSV
Mvan_0402|M.vanbaalenii_PYR-1 --MSNDRDFVEKRFSRPGEYRAAVIYTFVVVVLAAIAFAVYAFGD-----
Mflv_0340|M.gilvum_PYR-GCK --MTNDRDFVEKRFNRPGEYRSAVVYSLIVVALAAAAFVVYAFGP-----
MSMEG_0601|M.smegmatis_MC2_155 ---MIMTNDVEKRWRDPATFRSAVAYVLSVVAIAGVALIFYVM-------
TH_0798|M.thermoresistible__bu --VTDNVNDVEKRWHDPETFRAATRYVLTVVAIAMLALAVYVWIG-----
* : . . :
Mb3317c|M.bovis_AF2122/97 ---VDTAACGPPQRILLALGGPLILCAAGLWAFLRTYRVWRAEGTWWGWH
Rv3289c|M.tuberculosis_H37Rv ---VDTAACGPPQRILLALGGPLILCAAGLWAFLRTYRVWRAEGTWWGWH
MMAR_1244|M.marinum_M RAGVDTAACGMPERTLLALGAPAILFASGIWAFVRTYRVWRDGGTWWGWH
MUL_2643|M.ulcerans_Agy99 RAGVDTAACGMPERTLLALGAPAILFASGIWAFVRTYRVWRDGGTWWGWH
MAV_4261|M.avium_104 G-GVDTVACGPPQRTLLAFGGPLIALAGGIWAFVRTYRVWKARGTWWGWH
MAB_2521c|M.abscessus_ATCC_199 SVEVDKTLCGRHGTPAMAMAAPGVLMIGGIIAFVQTYRVWRRRGRWWVWQ
Mvan_0402|M.vanbaalenii_PYR-1 -----------RSSVFSAALVPVFLFAGGVGALLRTYRQWKGGGGWTAWQ
Mflv_0340|M.gilvum_PYR-GCK -----------RDSVFSAALVPAFLFAGGVGALIRTYREWKAGSGWTAWQ
MSMEG_0601|M.smegmatis_MC2_155 -----------DRTLLPAILVPTIMFVGGVGAFIRTYQVWRAEGTWPIWH
TH_0798|M.thermoresistible__bu -----------DRNIIAASTVPTILLVGGIGAFIKTYQVWKARGTWVMWQ
* * . .*: *:::**: *: . * *:
Mb3317c|M.bovis_AF2122/97 GAGWFLLTLMVLTLCIGVPPIAGPVMAP
Rv3289c|M.tuberculosis_H37Rv GAGWFLLTLMVLTLCIGVPPIAGPVMAP
MMAR_1244|M.marinum_M GAGWFLLTLMVVALSLGVPSIAGPVLAL
MUL_2643|M.ulcerans_Agy99 GAGWFLLTLMVVAVSLGVPSIAGPVLAL
MAV_4261|M.avium_104 GAGWFLLTVMALMVAMGVAPIAGPVLAL
MAB_2521c|M.abscessus_ATCC_199 GAGWFLLLLMVVTLML-----SGRMMT-
Mvan_0402|M.vanbaalenii_PYR-1 GAGWFLLLLMLLTLAVPGGAAFSN----
Mflv_0340|M.gilvum_PYR-GCK GAGWFLLLLMLLTLAVPGSAAFAG----
MSMEG_0601|M.smegmatis_MC2_155 GAGWFLLALSLFCLAFPGAALTS-----
TH_0798|M.thermoresistible__bu GAGWFLMAFMLVCLAVPGSAG-------
******: . . : .