For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGRHRAAARRPVRGARDRRRGTQARPLRQRRRPAPSGGVRRLRGVLARTAGHGRLYLMSRNRIQIDRRTA RWSDDPVTVADAMDFWSFAAGAANVIMQLSRPAVGYGVVESKVDSGNLSKHPWKRARTTFQYLAVAIFGS DDDRAAFREAVDGVHRQVKSGPSSPVRYNAFDRDLQMWVAACLFVGLEDTYQLLRGEMTHEQSEQFYLSA ATLGTTLQVSAEQWPLTRTDFDRYWDTACQQVEMDDVVRSYLTDLINLRMINPVLALPFRPLLKFLTAGF LAPVFRDALGVTWGGGRQWCFERLFLVVAFVNRFIPGFIRQGGSHVLLADVRMRVRRQRRLV
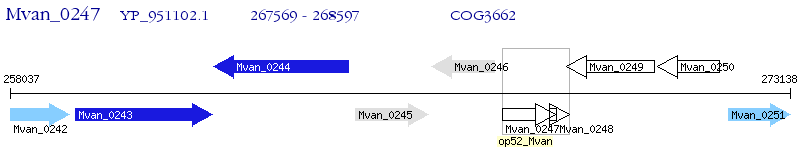
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0247 | - | - | 100% (342) | hypothetical protein Mvan_0247 |
| M. vanbaalenii PYR-1 | Mvan_0410 | - | 5e-61 | 45.86% (266) | hypothetical protein Mvan_0410 |
| M. vanbaalenii PYR-1 | Mvan_0854 | - | 1e-10 | 22.31% (260) | hypothetical protein Mvan_0854 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0282 | - | 6e-66 | 46.67% (285) | hypothetical protein Mb0282 |
| M. gilvum PYR-GCK | Mflv_0423 | - | 1e-142 | 86.32% (285) | hypothetical protein Mflv_0423 |
| M. tuberculosis H37Rv | Rv0276 | - | 6e-66 | 46.67% (285) | hypothetical protein Rv0276 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4329c | - | 8e-57 | 42.28% (272) | hypothetical protein MAB_4329c |
| M. marinum M | MMAR_0537 | - | 1e-63 | 47.86% (257) | hypothetical protein MMAR_0537 |
| M. avium 104 | MAV_4880 | - | 1e-62 | 49.03% (257) | hypothetical protein MAV_4880 |
| M. smegmatis MC2 155 | MSMEG_3590 | - | 1e-122 | 75.99% (279) | hypothetical protein MSMEG_3590 |
| M. thermoresistible (build 8) | TH_4430 | - | 1e-121 | 74.73% (281) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1201 | - | 3e-63 | 47.47% (257) | hypothetical protein MUL_1201 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0282|M.bovis_AF2122/97 ------------MAISLVAHQ---------------------------PI
Rv0276|M.tuberculosis_H37Rv ------------MAISLVAHQ---------------------------PI
MMAR_0537|M.marinum_M ------------MTIQEAAGR---------------------------LA
MUL_1201|M.ulcerans_Agy99 ------------MTIQEAAGR---------------------------LA
MAV_4880|M.avium_104 ------------MAIHEPVHQP---------------TAHVEPTGHVEPT
MAB_4329c|M.abscessus_ATCC_199 ------------MVTIEQVSEA---------------EAQLAGDGGPGTR
Mvan_0247|M.vanbaalenii_PYR-1 MGRHRAAARRPVRGARDRRRGTQARPLRQRRRPAPSGGVRRLRGVLARTA
Mflv_0423|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3590|M.smegmatis_MC2_155 ----------------------------------------------MTVE
TH_4430|M.thermoresistible__bu ------------MSLFDPHRASDPHP--------------------ASGS
Mb0282|M.bovis_AF2122/97 PHVERPMA--DPPRLQLARRRRSAAGPGGNEDSLMGVALLAGPANVIMEL
Rv0276|M.tuberculosis_H37Rv PHVERPMA--DPPRLQLARRRRSAAGPGGNEDSLMGVALLAGPANVIMEL
MMAR_0537|M.marinum_M PHVERPIS--DPPR-QVRRRKR-----GGFGDGLMGVALLAGPANVIMEL
MUL_1201|M.ulcerans_Agy99 PHVERPIS--DPPR-QVRQRKR-----GGFGNGLMGVALLAGPANVIMEL
MAV_4880|M.avium_104 SHVERRVT--DPPRPRRRKPLR----TLGIDEGLMGVALLAGPANVIMQL
MAB_4329c|M.abscessus_ATCC_199 SRRNEPVPPAAPLSPWWSWRRR-----PTVADNFMGLALLGGPANIIMQL
Mvan_0247|M.vanbaalenii_PYR-1 GHGRLYLMSRNRIQIDRRTARWS-DDPVTVADAMDFWSFAAGAANVIMQL
Mflv_0423|M.gilvum_PYR-GCK -------MNANRIQITRRTARWS-DDPVRVGDAMDFWSFAAGAANVIMQL
MSMEG_3590|M.smegmatis_MC2_155 G------TTHTRISVTRRTARWDPDESVTVADAMDFWAFAAGAANVIMQL
TH_4430|M.thermoresistible__bu G----LTVTDTRIHITRRTARWS-DAPVTVADAMDFWAFAAGAANVIMQL
: : :: .*.**:**:*
Mb0282|M.bovis_AF2122/97 AMPGVGYGVLESRVESGRLDRHPIKRARTTFTYVAVAVAGSDDQKAAFRR
Rv0276|M.tuberculosis_H37Rv AMPGVGYGVLESRVESGRLDRHPIKRARTTFTYVAVAVAGSDDQKAAFRR
MMAR_0537|M.marinum_M ARPGVGHGVMESRVESGRIDRHPIKRARTTFTYVAVASAGTDEQKAAFRR
MUL_1201|M.ulcerans_Agy99 ARPGVGHGVMESRVESGRIDRHPIKRARTTFTYVAVASAGTDEQKAAFRR
MAV_4880|M.avium_104 ANPGVGYGVMESRVESGRVDLHPIKRARTTFTYLAVATSGSEAQKDAFRR
MAB_4329c|M.abscessus_ATCC_199 ARPGVGYGVVDSKVESGRADLHPIKRARTTFTFLAVATLGTPEQKAAFRK
Mvan_0247|M.vanbaalenii_PYR-1 SRPAVGYGVVESKVDSGNLSKHPWKRARTTFQYLAVAIFGSDDDRAAFRE
Mflv_0423|M.gilvum_PYR-GCK SRPGVGYGVVESKVDSGNLMKHPWKRARTTFQYLAVAIFGSDDDRAAFRA
MSMEG_3590|M.smegmatis_MC2_155 ARPGVGHGVVESPVDSGNLLKHPWKRARTTFSYLAVAILGNDEDRAAFRT
TH_4430|M.thermoresistible__bu SRPEVAYGVMESRVDSGNLMKHPWKRARTTFQYLAVAVFGSESDRAMFRA
: * *.:**::* *:**. ** ******* ::*** *. :: **
Mb0282|M.bovis_AF2122/97 AVNKVHAQVYSTPESPVSYHAFDPELQLWVAACLYKGGVDVYRTFVGEMD
Rv0276|M.tuberculosis_H37Rv AVNKVHAQVYSTPESPVSYHAFDPELQLWVAACLYKGGVDVYRTFVGEMD
MMAR_0537|M.marinum_M AVNRVHAQVYSTPESPVSYSAFDVELQLWVAACLYKGGVDIYRTFVGEMD
MUL_1201|M.ulcerans_Agy99 AVNRVHAQVYSTPESPVSYSAFDVELQLWVAACLYKGGVDIYRTFVGEMD
MAV_4880|M.avium_104 AVNRAHAQVYSTPESPVSYNAFDPNLQLWVGACLYKGGLDIYRMFIGELD
MAB_4329c|M.abscessus_ATCC_199 ATNRAHAQVHSQPGDKVQYNAFDPELQKWVAVCLYKGFVDVYEAFVGPLT
Mvan_0247|M.vanbaalenii_PYR-1 AVDGVHRQVKSGPSSPVRYNAFDRDLQMWVAACLFVGLEDTYQLLRGEMT
Mflv_0423|M.gilvum_PYR-GCK AVDEAHRHVKSTPSSPVRYNAFDRDLQMWVAACLFVGLEDTYQLLRGEMT
MSMEG_3590|M.smegmatis_MC2_155 AVDGAHRQVRSGPQSPVRYNAFDRDLQMWVAACLFVGLEDTYQLLRGAMT
TH_4430|M.thermoresistible__bu AVDGVHRHVVSDRRSPVRYNAFDRELQMWVAACLFVGLEDSYRLLRGEMT
*.: .* :* * . * * *** :** **..**: * * *. : * :
Mb0282|M.bovis_AF2122/97 DEEADHHYRAGMAMGTTLQVPPQMWPPDRAAFDRYWRQSLDRVHIDDVVR
Rv0276|M.tuberculosis_H37Rv DEEADHHYRAGMAMGTTLQVPPQMWPPDRAAFDRYWRQSLDRVHIDDVVR
MMAR_0537|M.marinum_M DEEADRHYRDGMSMGTTLQVPAELWPPDRAAFDRYWQESLQKIHIDDAVR
MUL_1201|M.ulcerans_Agy99 DEEADRHYRDGMSMGTTLQVPAELWPPDRAAFDRYWQESLQKIHIDDAVR
MAV_4880|M.avium_104 GDDADRHYREGMTLATTLQVPPAMWPPDRAAFDRYWEESLAKVHIDDAVR
MAB_4329c|M.abscessus_ATCC_199 GETAERCLQEGAVMGTTLQMPLAMWPKSCAEFDEYWAQSMDLVHIDDHVR
Mvan_0247|M.vanbaalenii_PYR-1 HEQSEQFYLSAATLGTTLQVSAEQWPLTRTDFDRYWDTACQQVEMDDVVR
Mflv_0423|M.gilvum_PYR-GCK AEQADHFYASAWTLGTTLQVTEDQWPPTRADFDRYWDEACGQVAMDDVVR
MSMEG_3590|M.smegmatis_MC2_155 PEQAEQFYASAATLGTTLQVSPDQWPATRAGFDDYWNAACGQVAVDDVVA
TH_4430|M.thermoresistible__bu PGQAEEFYRSARPLGTTLQVAESQWPPTRAAFDEYWNNACARVVVDDRAR
::. . :.****:. ** : ** ** : : :** .
Mb0282|M.bovis_AF2122/97 DYLYPIVALRIRGIALPGPLRRLSEGIALLITTGFLPQRFRDEMRLPWDA
Rv0276|M.tuberculosis_H37Rv DYLYPIVALRIRGIALPGPLRRLSEGIALLITTGFLPQRFRDEMRLPWDA
MMAR_0537|M.marinum_M EYLYPIAASRVRGVALPGPLRRWSEGLALLITTGFLPQRFRDEMRLPWDA
MUL_1201|M.ulcerans_Agy99 EYLYPIAASRVRGVALPGPLRRWSEGLALLITTGFLPQRFRDEMRLPWDA
MAV_4880|M.avium_104 EYLYPIAANRIRGVWLPGPVRRLTEGFALFITTGFLPQRFRDEMRLPWDS
MAB_4329c|M.abscessus_ATCC_199 PYLYRIAS---ASIAVPPFMRGPVGAFSRFITTGFLPQRFRDEMGFAWDP
Mvan_0247|M.vanbaalenii_PYR-1 SYLTDLINLRMINPVLALPFR----PLLKFLTAGFLAPVFRDALGVTWGG
Mflv_0423|M.gilvum_PYR-GCK SYLLDLVDLRMINPVLGIPFR----PLLKFLTAGFLAPVFRDALGLTWGD
MSMEG_3590|M.smegmatis_MC2_155 TYLRDLVHLKMINPLLALPFR----PLLAFLTTGFLAPVFREVLGLKWGR
TH_4430|M.thermoresistible__bu AYLSDLLDLTMITPVLGRPFR----PLLRFLTVGFLAPVFRDALGVRWSD
** : : .* : ::*.***. **: : . *.
Mb0282|M.bovis_AF2122/97 TKQRRFDALMAVLRTVNRLMPRFVREFPFNLMLWDLDRRMRRGRPLV
Rv0276|M.tuberculosis_H37Rv TKQRRFDALMAVLRTVNRLMPRFVREFPFNLMLWDLDRRMRRGRPLV
MMAR_0537|M.marinum_M AKQRRFDRLIAVVGALNRMMPRIVREFPFNLMLWDLDRRIRTGRRLV
MUL_1201|M.ulcerans_Agy99 AKQRRFDRLIAMVGALNRMMPRIVREFPFNFMLWDLDRRIRTGRRLV
MAV_4880|M.avium_104 TKQRRFDRLIAVLATVNRYLPRFIRQFPFNVLLYDLDRRIRKGRPLV
MAB_4329c|M.abscessus_ATCC_199 SRERMFRWLLAVLGLAARLLPNGLRRFPFNALMLDLDWRIRTGRPLV
Mvan_0247|M.vanbaalenii_PYR-1 GRQWCFERLFLVVAFVNRFIPGFIRQGGSHVLLADVRMRVRRQRRLV
Mflv_0423|M.gilvum_PYR-GCK GRQRLFERLFLCVAFVNRFIPGFLRQGGSHVLLADVRMRVRRHRQLV
MSMEG_3590|M.smegmatis_MC2_155 GRQRLFEWLFLLVALGNRFLPIFVRQGTSYLLLADVRRRARAGLPLI
TH_4430|M.thermoresistible__bu RRQRRFEQLFLLVAFVNRFLPPFVRQGGSHVLLADVRRRARRGKRVI
:: * *: : * :* :*. :: *: * * ::