For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHLLEKFALAAAGAAGAVMLTATPAGAAPDPCAASEVARTVAEVATYTGNYLEANPEANRAITTISQQQG GPQSVAALKTYFDANPKVAADLQRLQQPLAALSGKCKLPLTLPQVFGLMQAAQQVPALPAAQNVGVTATR
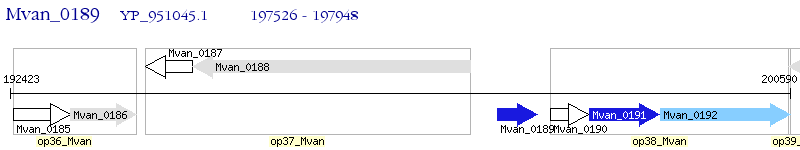
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0189 | - | - | 100% (140) | Fis family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5232 | - | 2e-14 | 39.81% (108) | hypothetical protein Mvan_5232 |
| M. vanbaalenii PYR-1 | Mvan_1639 | - | 2e-12 | 38.78% (98) | hypothetical protein Mvan_1639 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0209 | - | 5e-30 | 56.03% (116) | hypothetical protein Mb0209 |
| M. gilvum PYR-GCK | Mflv_0463 | - | 5e-48 | 76.38% (127) | Fis family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0203 | - | 5e-30 | 56.03% (116) | hypothetical protein Rv0203 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4528c | - | 2e-15 | 41.91% (136) | hypothetical protein MAB_4528c |
| M. marinum M | MMAR_0443 | - | 1e-31 | 51.49% (134) | exported protein |
| M. avium 104 | MAV_4971 | - | 2e-33 | 56.35% (126) | hypothetical protein MAV_4971 |
| M. smegmatis MC2 155 | MSMEG_0242 | - | 1e-38 | 63.56% (118) | hypothetical protein MSMEG_0242 |
| M. thermoresistible (build 8) | TH_3768 | - | 1e-09 | 33.02% (106) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1093 | - | 1e-31 | 51.49% (134) | exported protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0189|M.vanbaalenii_PYR-1 ------------------------------MHLLEK-----------FAL
Mflv_0463|M.gilvum_PYR-GCK --------------------------MIFAMNSAIN-----------TAL
MSMEG_0242|M.smegmatis_MC2_155 --------------------------MKAAITGAISGAKHDLRRTLGTVF
Mb0209|M.bovis_AF2122/97 --------------------------MKTGTATTRRR--------LLAVL
Rv0203|M.tuberculosis_H37Rv --------------------------MKTGTATTRRR--------LLAVL
MMAR_0443|M.marinum_M --------------------------MKSGNATKRRG--------LFAAL
MUL_1093|M.ulcerans_Agy99 --------------------------MKSGNATKRRG--------LFAAL
MAV_4971|M.avium_104 --------------------------MTSASAGRRRR--------IFAGL
MAB_4528c|M.abscessus_ATCC_199 MSATTSRRAAYGLLSAGVLAGGLALSALAPIAAAEPVPPTPAPAPAAPAA
TH_3768|M.thermoresistible__bu --------------------------MVLSARTVRRA---------VAGT
Mvan_0189|M.vanbaalenii_PYR-1 AAAGAAGAVML--TATPAGAAPDP----CAASEVARTVAEVATYTGNYLE
Mflv_0463|M.gilvum_PYR-GCK GVAALAAALIVGPVLPSASAAPDP----CAASEVARTVAEVATYTGNYLE
MSMEG_0242|M.smegmatis_MC2_155 VATAVGGAATAALLGPSATAATDP----CAASEVARTVGKVATDTGDYLD
Mb0209|M.bovis_AF2122/97 IALALPGAAVALLAEPSATGASDP----CAASEVARTVGSVAKSMGDYLD
Rv0203|M.tuberculosis_H37Rv IALALPGAAVALLAEPSATGASDP----CAASEVARTVGSVAKSMGDYLD
MMAR_0443|M.marinum_M VAVGLPGLALVTVAEPTATGATDP----CAASEVARTIGSVAKQTGDYLD
MUL_1093|M.ulcerans_Agy99 VAVGLPGLALVTVAEPTATGATDP----CAASEVARTIGSVAKQTGDYLD
MAV_4971|M.avium_104 IAAALPGAAVAVLAGPPATGANDP----CAASEVARTIGSVSKSMGDYLD
MAB_4528c|M.abscessus_ATCC_199 PAPVANAAAPAAATVTPATPTPDG----CQASKMTKTVGNVVQNAASYLE
TH_3768|M.thermoresistible__bu IGAGAIAGATLFSSIPFAFADPEPEPPGCTAADLAGVAAGVSAATSAYLF
. . * : * *:.:: . . * . **
Mvan_0189|M.vanbaalenii_PYR-1 ANPEANRAITTISQQQGGPQSVAALKTYFDANPKVAADLQRLQQPLAALS
Mflv_0463|M.gilvum_PYR-GCK ANPKANTALTSISQQ-GGPESVAALKTYFDANPKVASDLQRLQAPLVTLS
MSMEG_0242|M.smegmatis_MC2_155 ENPQTNQALTTISKQQAGPATLASIKTYLDANPQVAKDLQRLQQPLTALG
Mb0209|M.bovis_AF2122/97 SHPETNQVMTAVLQQQVGPGSVASLKAHFEANPKVASDLHALSQPLTDLS
Rv0203|M.tuberculosis_H37Rv SHPETNQVMTAVLQQQVGPGSVASLKAHFEANPKVASDLHALSQPLTDLS
MMAR_0443|M.marinum_M SHPETNQAMTAAFQGSAGPQSFGSLKSYLEKNPKVASDLQSLSQPLTNLS
MUL_1093|M.ulcerans_Agy99 SHPETNQAMTAAFQGSAGPQSFGSLKSYLEKNPKVASDLQSLSQPLTNLS
MAV_4971|M.avium_104 AHPETNQAMTSMLQQQAGPQSVNGLKSYFDANPKVAGDMTSIAQPLTSLS
MAB_4528c|M.abscessus_ATCC_199 QHPDVDAAFTEIKAAPQDQIATK-TQSYLVSRPDVAASLGGIAAPLNELR
TH_3768|M.thermoresistible__bu THPEVNDFFTG-LEGQHRADVRDTVAEYLDANPQVKAELTHIRQPLVDIK
:*..: :* :: .*.* .: : ** :
Mvan_0189|M.vanbaalenii_PYR-1 GKCKLPLTLPQVFG---------------LMQAAQQ--------------
Mflv_0463|M.gilvum_PYR-GCK GTCRLPVTLPQIFG---------------LMQAAQQ--------------
MSMEG_0242|M.smegmatis_MC2_155 TKCQLPVTLPQLLG---------------LVQAAQQQGTAAAGSLPGGLT
Mb0209|M.bovis_AF2122/97 TRCSLPISGLQAIG---------------LMQAVQG--------------
Rv0203|M.tuberculosis_H37Rv TRCSLPISGLQAIG---------------LMQAVQG--------------
MMAR_0443|M.marinum_M TQCRLPITLSQAVG---------------LAQAAQDGG------------
MUL_1093|M.ulcerans_Agy99 TQCRLPITLSQAVG---------------LAQAAQDGG------------
MAV_4971|M.avium_104 LQCKLPISIPQALG---------------MVQQAQGAAGGL---------
MAB_4528c|M.abscessus_ATCC_199 TKCGLPLDLAQVVNGGGLNPAAMGVNPALLTALSQNPAAQQVAQNPAAQA
TH_3768|M.thermoresistible__bu NRCGGPDLATDDIP------------------------------------
* * : .
Mvan_0189|M.vanbaalenii_PYR-1 -----------VPAL----------PAAQNVG------VTATR-------
Mflv_0463|M.gilvum_PYR-GCK -----------GPAQ----------PVAHSV-------APAPR-------
MSMEG_0242|M.smegmatis_MC2_155 SPQIAGVPGVVGPAQRT-----PISPVSQGSGPLPGPAVTSTR-------
Mb0209|M.bovis_AF2122/97 ---------------------------------------ARR--------
Rv0203|M.tuberculosis_H37Rv ---------------------------------------ARR--------
MMAR_0443|M.marinum_M --ALSGLPAVGLPL--------TPGNLPVAAGPLPGPAPANRAG------
MUL_1093|M.ulcerans_Agy99 --ALSGLPAVGLPL--------TPGNLPVAAGPLPGPARANRAG------
MAV_4971|M.avium_104 -PALPGNPAGASPLGGAAGPAGTPPAVPQPAGPLSGPPRGNTVG------
MAB_4528c|M.abscessus_ATCC_199 LVQNPAVQAAIQNPAPAATFPQGPGPAPGPAQPVQPAAPAGPVGTGPAPG
TH_3768|M.thermoresistible__bu --------------------------------------------------
Mvan_0189|M.vanbaalenii_PYR-1 ---
Mflv_0463|M.gilvum_PYR-GCK ---
MSMEG_0242|M.smegmatis_MC2_155 ---
Mb0209|M.bovis_AF2122/97 ---
Rv0203|M.tuberculosis_H37Rv ---
MMAR_0443|M.marinum_M ---
MUL_1093|M.ulcerans_Agy99 ---
MAV_4971|M.avium_104 ---
MAB_4528c|M.abscessus_ATCC_199 PRV
TH_3768|M.thermoresistible__bu ---