For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQNDPRMRSESGVLGQIDAQQVPRYAGLGTFARLPQRHEVASHDIAVIGVPFDSGVTYRPGARFGPAAIR QASRLLKPYHPALEVSPFATAQVVDAGDIAANPFDITTAVEQIKDGILGLVTSPEQRFVLLGGDHTIALP ALQAVNELHGPVALVHFDAHLDTWDTYFGAPCTHGTPFRRASEQGLIVKGHSAHVGIRGSLYDAADLLDD AELGFTAVHCRDIDRIGVDGVIERVLERVGEHPVYVSIDVDVLDPAFAPGTGTPEIGGMTSRELVAVLRA MRALNIVGADVVEVAPAYDHAEVTAVAAANLAYELISLMVG
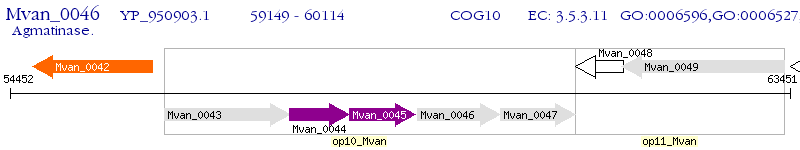
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0046 | - | - | 100% (321) | putative agmatinase |
| M. vanbaalenii PYR-1 | Mvan_0161 | - | e-108 | 60.78% (306) | putative agmatinase |
| M. vanbaalenii PYR-1 | Mvan_3024 | - | 1e-49 | 38.14% (333) | putative agmatinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0797 | - | 1e-176 | 94.70% (321) | putative agmatinase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1072 | speB | 1e-154 | 81.82% (319) | agmatinase |
| M. thermoresistible (build 8) | TH_3257 | - | 4e-42 | 34.77% (325) | PUTATIVE putative agmatinase |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0046|M.vanbaalenii_PYR-1 ----------------MQNDPRMRSESGVLGQIDAQQVPRYAGLGTFARL
Mflv_0797|M.gilvum_PYR-GCK MQRRFDAGTGMSEESSVQNDPRMQSASGVLGQIDAQQVPRYAGFGTFARL
MSMEG_1072|M.smegmatis_MC2_155 ----------------MEGQPYVRSASGKVGQVNATEVPRYAGIATFARL
TH_3257|M.thermoresistible__bu -----------VWRQPVDRSRDPHREPGPINLQRHQFTPAYSGIATLFGL
:: . : .* :. .* *:*:.*: *
Mvan_0046|M.vanbaalenii_PYR-1 P-----QRHEVASHDIAVIGVPFDSGVTYRPGARFGPAAIRQASRLLKP-
Mflv_0797|M.gilvum_PYR-GCK P-----QRHEVPGHDIAVIGVPFDSGVTYRPGARFGPAAIRQASRLLKP-
MSMEG_1072|M.smegmatis_MC2_155 P-----QRFEVLDHDIAVVGVPFDSGVTYRPGARFGPSAIREASRLLKP-
TH_3257|M.thermoresistible__bu PLCLNQDDLRAGAVDVAVVGAPVDVSLGHR-GAAYGPRAIRADERVLPHT
* : .. *:**:*.*.* .: :* ** :** *** .*:*
Mvan_0046|M.vanbaalenii_PYR-1 ----YHPALEVSPFATAQVVDAGDIAANPFDITTAVEQIKDGILGLVTSP
Mflv_0797|M.gilvum_PYR-GCK ----YNPALEVAPFATAQVVDAGDIAANPFDITEAVDQIKDGILGLVTRP
MSMEG_1072|M.smegmatis_MC2_155 ----YHPALDVSPFAQAQVVDAGDLGVNPFNIDEAVGQIRDGVTELLDRP
TH_3257|M.thermoresistible__bu PTMLINDSTRVKPFEVLNVVDYGDAAVDPLSIENSMGPIRELVRDVAEVG
: : * ** :*** ** ..:*:.* :: *:: : :
Mvan_0046|M.vanbaalenii_PYR-1 EQRFVLLGGDHTIALPALQAVNELHGP--VALVHFDAHLDTWDTYFGAPC
Mflv_0797|M.gilvum_PYR-GCK EQRFVLLGGDHTIALPALQAVNELHGP--VALVHFDAHLDTWDTYFGAPC
MSMEG_1072|M.smegmatis_MC2_155 DQRVVLLGGDHTIALPALQAMHAIHGP--VALVHFDAHLDTWDTYFGAPC
TH_3257|M.thermoresistible__bu -AVPLVLGGDHSILWPNAAAIADVYGAGKVGVVHFDAHPDCADDVVGHHA
::*****:* * *: ::*. *.:****** * * .* .
Mvan_0046|M.vanbaalenii_PYR-1 THGTPFRRASEQGLIVKGHSAHVGIRGSLYDAADLLDDA-ELGFTAVHCR
Mflv_0797|M.gilvum_PYR-GCK THGTPFRRASEQGLIVKGHSAHVGIRGSLYDSADLLDDA-ELGFTAVRCR
MSMEG_1072|M.smegmatis_MC2_155 THGTPFRRASEQGLIVKGHSAHVGIRGSLYDRADLLDDE-SLGFTVVHCR
TH_3257|M.thermoresistible__bu SHGTPIRRLIDDEHIPGRNFIQVGLRSAIAPDEELFAWMRDNGLRTHFMA
:****:** :: * : :**:*.:: :*: . *: .
Mvan_0046|M.vanbaalenii_PYR-1 DIDRIGVDGVIERVLERVGEHP--VYVSIDVDVLDPAFAPGTGTPEIGGM
Mflv_0797|M.gilvum_PYR-GCK DIDRIGVDGVIERVLERVGDHP--VYVSIDIDVLDPAFAPGTGTPEIGGM
MSMEG_1072|M.smegmatis_MC2_155 DIDRIGVDGVISRIHERVGEYP--VYVSIDIDVLDPAFAPGTGTPEIGGM
TH_3257|M.thermoresistible__bu EIDRRGFHTVLQDAIDEALDGPEYLYLSIDIDVLDPAYAPGTGTPEPPGL
:*** *.. *:. :.. : * :*:***:******:******** *:
Mvan_0046|M.vanbaalenii_PYR-1 TSRELVAVLRAMRALN-IVGADVVEVAPAYDHAEVTAVAAANLAYELIS-
Mflv_0797|M.gilvum_PYR-GCK TSRELVAVLRAMRALD-IVGADVVEVAPSYDHAEVTAVAAANLAYELIS-
MSMEG_1072|M.smegmatis_MC2_155 TSRELVAVLRAMRGLN-IVGADIVEVAPAYDSGDVTAVAAANLAYELIT-
TH_3257|M.thermoresistible__bu TNRELLPAIRRICHETPVVGLEIVEVAPHLDPGYTTTMNARRVIFEALTG
*.***:..:* : :** ::***** * . .*:: * .: :* ::
Mvan_0046|M.vanbaalenii_PYR-1 LMVG----------------------
Mflv_0797|M.gilvum_PYR-GCK LMVG----------------------
MSMEG_1072|M.smegmatis_MC2_155 LMADGF--------------------
TH_3257|M.thermoresistible__bu LAMRRLGLPGPDYLHPDVAGPPSNRV
*