For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDFSEAEWESLALETLAQQEWLPLNGSAIAPGTENGRASWDELVLPDRMLAKLRELNAHVPGEYLEQAR AAILQPSSQDAIAENYRLHQYLVGGYRGISYVDSDGIEQNPTIRLISHRPEENELLAVQQVTIRDAEHDR RFDIVLYLNGMPIAFFELKQAGSKYADLPGAHAQFATYLREFPMAFRFAVLNVISDGLTARYGTPFTPLE HFAPWNVDDDGKPVAFGDPVDDVHLGTELEYLIDGLFNPERFLQLVRNFTAFDAGADGLIKRIAKPHQYF AVTKAVGSTVTAAESNGKAGVVWHTQGSGKSMEMELYTHLVAQQPKLKNPTVVVVTDRKDLDGQLYATFD RSKLLGESPVKVTTRSQLRDELSNRTTGGIYFTTLQKFGLSKAERESGADHPLLTDRRNIIVVVDEAHRS HYDDLDGYARHIRDALPNAVYIAFTGTPISEADRDTRDVFGPDIDVYDLTRAVNDKATVPVFFEPRLIKV ALAQGVTEDDLDKAADEVTAGLDDVERDQIEKSVAVINAVYGAPDRLAALARDIVDHWEVRSQEMRKFIS CPGKAFIVGATREICAELYEEIVKLKPEWHDDAVDKGVIKVVYSGSAKDQGLVAKHVRRDGQNKTIQQRL RDPDDELQIVLVKDMLLTGFDAPPLHTLYLDRPLKGALLMQTLARVNRTFREKPNGLLVAYAPLVENLNK ALAEYTQTDRTEKPVGKNIDEAVALTETLIAQLDALCAGYDWRAKVAQPHGWMKAAVGLTNYLRSPATAG NQVAEGEATVSDRFRALANQLSRAWSLCAGNQALDALRPTAKFYEEVRVWMAKFDANDRQASGKPVPEAI RRMLESLVYDSTASDGIVDIYDAAGLPKPSLSDLTPEFEAKAASASNPHLAIEALRAVITEEAVRATKSN VVRQRAFSERLTDLMRRYTNQQLTSAEVIAELIEMAKEVAAESNRGAHFSPPLSHDELAFYDAVAQNESA VELQGEDVLAQIARELVGVMQRDTKTDWTVRDDVRAKLRSSIKRLLVKYKYPPDKQPEAIKLVIEQMEAL APGYADAARAG
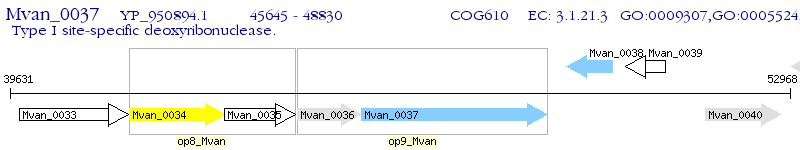
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0037 | - | - | 100% (1061) | HsdR family type I site-specific deoxyribonuclease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3649 | - | 1e-135 | 79.47% (302) | type I restriction-modification enzyme, R subunit |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0037|M.vanbaalenii_PYR-1 MTDFSEAEWESLALETLAQQEWLPLNGSAIAPGTENGRASWDELVLPDRM
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 LAKLRELNAHVPGEYLEQARAAILQPSSQDAIAENYRLHQYLVGGYRGIS
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 YVDSDGIEQNPTIRLISHRPEENELLAVQQVTIRDAEHDRRFDIVLYLNG
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 MPIAFFELKQAGSKYADLPGAHAQFATYLREFPMAFRFAVLNVISDGLTA
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 RYGTPFTPLEHFAPWNVDDDGKPVAFGDPVDDVHLGTELEYLIDGLFNPE
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 RFLQLVRNFTAFDAGADGLIKRIAKPHQYFAVTKAVGSTVTAAESNGKAG
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 VVWHTQGSGKSMEMELYTHLVAQQPKLKNPTVVVVTDRKDLDGQLYATFD
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 RSKLLGESPVKVTTRSQLRDELSNRTTGGIYFTTLQKFGLSKAERESGAD
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 HPLLTDRRNIIVVVDEAHRSHYDDLDGYARHIRDALPNAVYIAFTGTPIS
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 EADRDTRDVFGPDIDVYDLTRAVNDKATVPVFFEPRLIKVALAQGVTEDD
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 LDKAADEVTAGLDDVERDQIEKSVAVINAVYGAPDRLAALARDIVDHWEV
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 RSQEMRKFISCPGKAFIVGATREICAELYEEIVKLKPEWHDDAVDKGVIK
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 VVYSGSAKDQGLVAKHVRRDGQNKTIQQRLRDPDDELQIVLVKDMLLTGF
MAV_3649|M.avium_104 --------------------------------------------------
Mvan_0037|M.vanbaalenii_PYR-1 DAPPLHTLYLDRPLKGALLMQTLARVNRTFREKPNGLLVAYAPLVENLNK
MAV_3649|M.avium_104 -------------------------------MSPGGGIVG----------
.*.* :*.
Mvan_0037|M.vanbaalenii_PYR-1 ALAEYTQTDRTEKPVGKNIDEAVALTETLIAQLDALCAGYDWRAKVAQPH
MAV_3649|M.avium_104 -------------------------------------AVSTRRLSVAQAR
* * .***.:
Mvan_0037|M.vanbaalenii_PYR-1 GWMKAAVGLTNYLRSPATAGNQVAEGEATVSDRFRALANQLSRAWSLCAG
MAV_3649|M.avium_104 -----RIAVAAYLRSPSTPGNQVPEGEPTLGDRFRNQANHLARAWSLCVG
:.:: *****:*.****.***.*:.**** **:*:******.*
Mvan_0037|M.vanbaalenii_PYR-1 NQALDALRPTAKFYEEVRVWMAKFDANDRQASGKPVPEAIRRMLESLVYD
MAV_3649|M.avium_104 NETLDTLRPTVKFYEQVRVWMGKFDAQQRQAEGKPIPDDIKRLLSSLVAE
*::**:****.****:*****.****::***.***:*: *:*:*.*** :
Mvan_0037|M.vanbaalenii_PYR-1 STASDGIVDIYDAAGLPKPSLSDLTPEFEAKAASASNPHLAIEALRAVIT
MAV_3649|M.avium_104 STASGEIVDIYDAAGLPKPSLSELGPDYEAKAKAAAHPHLAIEALRAVLT
****. ****************:* *::**** :*::***********:*
Mvan_0037|M.vanbaalenii_PYR-1 EEAVRATKSNVVRQRAFSERLTDLMRRYTNQQLTSAEVIAELIEMAKEVA
MAV_3649|M.avium_104 KELAAATKNNVVRQRAFSDRISDLMRRYTNQQLTSAEVIAELIAMAKDVA
:* . ***.*********:*::********************* ***:**
Mvan_0037|M.vanbaalenii_PYR-1 AESNRGAHFSPPLSHDELAFYDAVAQNESAVELQGEDVLAQIARELVGVM
MAV_3649|M.avium_104 AESDRGKHFTPPLSNDELAFYDAVAQNESAVELQGEDVLAQIARELVGVM
***:** **:****:***********************************
Mvan_0037|M.vanbaalenii_PYR-1 QRDTKTDWTVRDDVRAKLRSSIKRLLVKYKYPPDKQPEAIKLVIEQMEAL
MAV_3649|M.avium_104 QRDTKTDWTVRDDVRAKLRSSIKRLLVKYRYPPDKQPEAIKLVIEQMEAL
*****************************:********************
Mvan_0037|M.vanbaalenii_PYR-1 APGYAD-AARAG------
MAV_3649|M.avium_104 APEYAERAGRMAKGEDCE
** **: *.* .