For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTEEDDALNPPAHLEGLKGMDLVRRTLEEARGAARQQGKNVGLGRYSPTARRVAGSGRRRWSGPGPDSRD PQLLGAVTGDVARTRGWSAKVAEGAVFGRWRAVVGDQIAAHAAPTALHEGVLTVSAESTAWATQLRMVQS QILAKIAAAVGDGVVTSLKIVGPVGPSWRKGPYTVPGRGPRDTYG
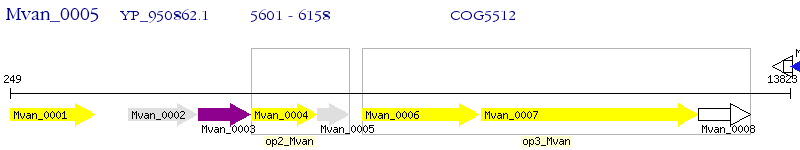
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0005 | - | - | 100% (185) | hypothetical protein Mvan_0005 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0004 | - | 6e-60 | 67.26% (168) | hypothetical protein Mb0004 |
| M. gilvum PYR-GCK | Mflv_0823 | - | 5e-85 | 81.62% (185) | hypothetical protein Mflv_0823 |
| M. tuberculosis H37Rv | Rv0004 | - | 6e-60 | 67.26% (168) | hypothetical protein Rv0004 |
| M. leprae Br4923 | MLBr_00004 | - | 2e-61 | 63.89% (180) | hypothetical protein MLBr_00004 |
| M. abscessus ATCC 19977 | MAB_0005 | - | 5e-77 | 75.57% (176) | hypothetical protein MAB_0005 |
| M. marinum M | MMAR_0004 | - | 3e-63 | 70.59% (170) | hypothetical protein MMAR_0004 |
| M. avium 104 | MAV_0004 | - | 4e-66 | 74.25% (167) | hypothetical protein MAV_0004 |
| M. smegmatis MC2 155 | MSMEG_0004 | - | 7e-79 | 81.36% (177) | hypothetical protein MSMEG_0004 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0004 | - | 1e-62 | 70.00% (170) | hypothetical protein MUL_0004 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0004|M.bovis_AF2122/97 MTGSV-----DRPDQNRG--ERLMKSPGLDLVRRTLDEARAAARARGQDA
Rv0004|M.tuberculosis_H37Rv MTGSV-----DRPDQNRG--ERSMKSPGLDLVRRTLDEARAAARARGQDA
MMAR_0004|M.marinum_M MSDDG-----EQPGPGDG--AARDELSGMDLVRRTLAEARAAARARGQDP
MUL_0004|M.ulcerans_Agy99 MNGDG-----EQPGPGDG--AARDELPSMDLVRRTLAEARAAARARGQDP
MAV_0004|M.avium_104 MSDD------QSPSPSG-------EPTAMDLVRRTLEEARAAARAQGKDA
MLBr_00004|M.leprae_Br4923 MIESN-----ESYSYGGDTIEPLGTLSGFDLVRRALEEARAAACAQGKDA
Mvan_0005|M.vanbaalenii_PYR-1 MT--------EEDD-ALNPPAHLEGLKGMDLVRRTLEEARGAARQQGKNV
Mflv_0823|M.gilvum_PYR-GCK MS--------EDDDNNIGPPPHLEGLKGMDLVRRALEEARGAARQQGKNV
MSMEG_0004|M.smegmatis_MC2_155 MTGPFDDDGPEEDAPVPAPPDHLAGLRGIDLVRRTLEEARGAARSQGKDV
MAB_0005|M.abscessus_ATCC_1997 MS--------EEEA---WPPEHLAGVKGMDLVRRVLEEARGAAKQQGKDI
* : .:*****.* ***.** :*::
Mb0004|M.bovis_AF2122/97 GRGRVAS-VASGRVAG--RRRSWSGPGPDIRDPQPLGKAARELAKKRGWS
Rv0004|M.tuberculosis_H37Rv GRGRVAS-VASGRVAG--RRRSWSGPGPDIRDPQPLGKAARELAKKRGWS
MMAR_0004|M.marinum_M GRGFAAG-PAPRRVAG--RRRSWSGPGPDTRDPQPLGKLTRDLAKKRGWS
MUL_0004|M.ulcerans_Agy99 GRGFAAG-PAPRRVAG--RRRSWSGPGPDTRDPQPLGKLTRDLAKKRGWS
MAV_0004|M.avium_104 GRGRAAA-PTPRRVAG--QRRSWSGPGPDARDPQPLGRLARDLARKRGWS
MLBr_00004|M.leprae_Br4923 GRGHVVP-PVPFRVTD--RRRNWSGPGPDVRDPQPLGKVAHDLAKKRGWS
Mvan_0005|M.vanbaalenii_PYR-1 GLGRYSP--TARRVAGSGRRR-WSGPGPDSRDPQLLGAVTGDVARTRGWS
Mflv_0823|M.gilvum_PYR-GCK GQGRTAPSGSPRRGTARSRRR-WSGPGPDNRDPQLLGSLTGDLARARGWS
MSMEG_0004|M.smegmatis_MC2_155 GRGRSGP---ARRVGGNRRRRTWSGPGPDARDPQLLGAVTQDLAKSRGWS
MAB_0005|M.abscessus_ATCC_1997 GRGGRSP--EQRRRVAGGRRT-WSGPGPDARDPQLLGRAAGDLAKRRGWS
* * * :* ******* **** ** : ::*: ****
Mb0004|M.bovis_AF2122/97 VRVAEGMVLGQWSAVVGHQIAEHARPTALNDGVLSVIAESTAWATQLRIM
Rv0004|M.tuberculosis_H37Rv VRVAEGMVLGQWSAVVGHQIAEHARPTALNDGVLSVIAESTAWATQLRIM
MMAR_0004|M.marinum_M GHVAEGTVLGQWSRVVGAQIADHATPTALNEGVLSVTAESTAWATQLRIM
MUL_0004|M.ulcerans_Agy99 GHVAEGTVLGQWSQVVGAQIADHATPTALNEGVLSVTAESTAWATQLRIM
MAV_0004|M.avium_104 AQVAEGTVLGNWTAVVGHQIADHAVPTGLRDGVLSVSAESTAWATQLRMM
MLBr_00004|M.leprae_Br4923 AQVAEGRVFGQWASMVGGQIADHAFPVGLNNGVLSVTAESTAWATQLRIM
Mvan_0005|M.vanbaalenii_PYR-1 AKVAEGAVFGRWRAVVGDQIAAHAAPTALHEGVLTVSAESTAWATQLRMV
Mflv_0823|M.gilvum_PYR-GCK GRVAQGAVFGRWRAVVGDQIADHASPTTLTEGVLTVSAESTAWATQLRMV
MSMEG_0004|M.smegmatis_MC2_155 ARVAEGSVIGRWRAVVGDQIADHATPTALNEGVLTVTAESTAWATQLRMV
MAB_0005|M.abscessus_ATCC_1997 SRVSEGAVFGRWEAVVGEQIAAHATPTALNEGVLTVAAESTAWATQLRLV
:*::* *:*.* :** *** ** *. * :***:* ***********::
Mb0004|M.bovis_AF2122/97 QAQLLAKIAAAVGNDVVRSLKITGPAAPSWRKGPRHIAGRGPRDTYG
Rv0004|M.tuberculosis_H37Rv QAQLLAKIAAAVGNDVVRSLKITGPAAPSWRKGPRHIAGRGPRDTYG
MMAR_0004|M.marinum_M QSQLLAKIAAAVGNGVVTSLKITGPASPSWRKGPRHIAGRGPRDTYG
MUL_0004|M.ulcerans_Agy99 QSQLLAKIAAAVGNGVVTSLKITGPASPSWRKGPRHIAGRGPRDTYG
MAV_0004|M.avium_104 QAQLLAKIAAAVGNGVVTSLKITGPAAPSWRKGPRHIAGRGPRDTYG
MLBr_00004|M.leprae_Br4923 QAQLLAKIAAAVGNGVVTSLKITGPTAPSWRKGPWHIAGRGPRDTYG
Mvan_0005|M.vanbaalenii_PYR-1 QSQILAKIAAAVGDGVVTSLKIVGPVGPSWRKGPYTVPGRGPRDTYG
Mflv_0823|M.gilvum_PYR-GCK QSQILAKIAAAVGDGVVTSLKIVGPVGPSWRKGPYNVRGRGPRDTYG
MSMEG_0004|M.smegmatis_MC2_155 QSQLLAKIAAVVGDGVVTTLKIVGPAGPSWRKGRYHVSGRGPRDTYG
MAB_0005|M.abscessus_ATCC_1997 QAQLLAKIAAAIGDGVVTSLKISGPTAPSWRKGPRHIAGRGPRDTYG
*:*:******.:*:.** :*** **..****** : *********