For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NAREYRDSSRKRLTDYPRPSVAVDSAVLTLDEQGGLVVLQVRRENRRGWGLPGTFLHPGERLSDAVKRSL RVKANVEGLDPRQLYVFDDPDRDDRGWVLSVAHVDVVRPERLADRFAESTRLMPVGSPGRLSFDHADIVA RALSYLRDRYARSPDPDHLLDDEFTIRELRLAHEAVAGEPLQRDWFRRAMEPQLVATGVMTSRGRGRPAE LFRRP*
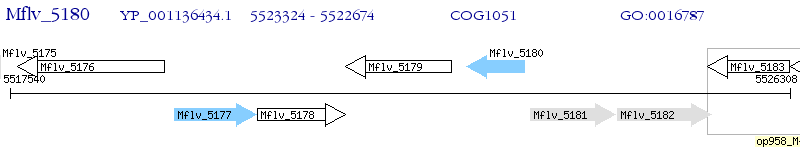
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5180 | - | - | 100% (216) | NUDIX hydrolase |
| M. gilvum PYR-GCK | Mflv_3620 | - | 2e-11 | 29.41% (170) | NUDIX hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1619c | - | 4e-11 | 29.41% (187) | hypothetical protein Mb1619c |
| M. tuberculosis H37Rv | Rv1593c | - | 4e-11 | 29.41% (187) | hypothetical protein Rv1593c |
| M. leprae Br4923 | MLBr_01224 | - | 2e-09 | 28.98% (176) | hypothetical protein MLBr_01224 |
| M. abscessus ATCC 19977 | MAB_2675 | - | 4e-09 | 30.51% (177) | hypothetical protein MAB_2675 |
| M. marinum M | MMAR_2390 | - | 4e-09 | 28.95% (190) | hypothetical protein MMAR_2390 |
| M. avium 104 | MAV_3192 | - | 8e-11 | 30.53% (190) | hydrolase, NUDIX family protein |
| M. smegmatis MC2 155 | MSMEG_1268 | - | 1e-81 | 69.30% (215) | nudix hydrolase |
| M. thermoresistible (build 8) | TH_0810 | - | 6e-12 | 31.68% (202) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1567 | - | 3e-09 | 28.95% (190) | hypothetical protein MUL_1567 |
| M. vanbaalenii PYR-1 | Mvan_1101 | - | 1e-85 | 73.02% (215) | NUDIX hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1619c|M.bovis_AF2122/97 ---------MAHGSTAHEVLAVVFQVRGVGMSRG----AAKPQLNVLLWQ
Rv1593c|M.tuberculosis_H37Rv ---------MAHGSTAHEVLAVVFQVRGVGMSRG----AAKPQLNVLLWQ
MLBr_01224|M.leprae_Br4923 ---------MAYASTAHEVLAVVFQVRRVAEQVKGTPQKAKPQLSVLLWE
MAV_3192|M.avium_104 ----------------------MFQVREASEPVTKGRHRAKPQLNVLLWQ
MMAR_2390|M.marinum_M ---------MPNSNTAHEVLAAVFQVRQITAASR----GGKPQLNVLLWQ
MUL_1567|M.ulcerans_Agy99 ---------MPNSNTAHEVLAAVFQVRHITAASR----RGKPQLNVLLWQ
MAB_2675|M.abscessus_ATCC_1997 ---------MVHTSTEHEVLAVVFQVRHFDKAQT-------PELAVLLWQ
TH_0810|M.thermoresistible__bu ------------------VLAVVFQVRGLDSRQ--------PALHVLLWQ
Mflv_5180|M.gilvum_PYR-GCK ---------MNAREYRDSSRKRLTDYPRPSVAVDSAVLTLDEQGGLVVLQ
Mvan_1101|M.vanbaalenii_PYR-1 MTTVASTPDMSERRYRDSSDKTLADYPRPSVAVDTAVLTLDEQQGLVVLQ
MSMEG_1268|M.smegmatis_MC2_155 ---------MDNAEYQDSSGRTLGDYPRPSVAVDAAVLTVGPDAGLSVLQ
: : : : :
Mb1619c|M.bovis_AF2122/97 RAKEPQRGAWSLPGGRLRNDEDMTSSVRRQLAEKVDLRELAHLEQLAVFS
Rv1593c|M.tuberculosis_H37Rv RAKEPQRGAWSLPGGRLRNDEDMTSSVRRQLAEKVDLRELAHLEQLAVFS
MLBr_01224|M.leprae_Br4923 RSQDPQRGAWSLPGGRLRNDEDMTYSVRRQLAEKVDLRELAHLEQLAVFS
MAV_3192|M.avium_104 RAKEPQRGAWSLPGGRLRNDEDMTTSVRRQLAEKVDLKELAHLEQLAVFS
MMAR_2390|M.marinum_M RAQDPQKGAWSLPGGRLRSDEDLTTSVRRQLAEKVDLRELSHLEQLAVFS
MUL_1567|M.ulcerans_Agy99 RAQDPQKGAWSLPGGRLRSDEDLTTSVRRQLAEKVDLRELSHLEQLAVFS
MAB_2675|M.abscessus_ATCC_1997 RALDPHRGAWALPGGRVRADEDLPSSIRRQLAEKVDVRELAHLEQLAVFS
TH_0810|M.thermoresistible__bu RALEPERGRWSLPGGLLGTDEDLTSSVRRQLAEKVDLRELAHLEQLAVFS
Mflv_5180|M.gilvum_PYR-GCK VRRENRRG-WGLPGTFLHPGERLSDAVKRSLRVKANVEGLD-PRQLYVFD
Mvan_1101|M.vanbaalenii_PYR-1 VRRPNADG-WALPGTFLHEGETLAMAVARSLQVKANVEGLL-PRQLHVFD
MSMEG_1268|M.smegmatis_MC2_155 VRRAQGRG-WALPGTFLHPGETLADAVNRALATKANVHGLR-PRQLHVFD
* *.*** : .* :. :: * * *.::. * .** **.
Mb1619c|M.bovis_AF2122/97 DPHRLPG-IRMIASTYLGVVPSP-ATPELPADTRWHPVSSLPPMAFDHGP
Rv1593c|M.tuberculosis_H37Rv DPHRLPG-IRMIASTYLGVVPSP-ATPELPADTRWHPVSSLPPMAFDHGP
MLBr_01224|M.leprae_Br4923 EPARLPG-TRMIASTFLGLVPSP-ATPELPPDTRWHPLNTLPSMAFDHGP
MAV_3192|M.avium_104 DPNRVPG-TRMIASTFLGLVPSP-ASPELPPDTRWHPVNCLPPMAFDHGQ
MMAR_2390|M.marinum_M DPHRVPGDTRVIASTFLGLVPSP-ATPELPADTRWHPVNALPPMAFDHGP
MUL_1567|M.ulcerans_Agy99 DPHRVPGDTRVIASTFLGLVPSP-ATPELPADTRWHPVNALPPMAFDHGP
MAB_2675|M.abscessus_ATCC_1997 EPRRVPG-PRTIASTFLGLVPFP-ATPALPDDTQWHRVNHLPAMAFDHQV
TH_0810|M.thermoresistible__bu DPHRVPG-VRTIASTYLGLVPSP-ASPALPDDTRWHPVHALPPMAFDHGP
Mflv_5180|M.gilvum_PYR-GCK DPDRDDR-GWVLSVAHVDVVRPERLADRFAESTRLMPVGSPGRLSFDHAD
Mvan_1101|M.vanbaalenii_PYR-1 ALDRDPR-GWVLSVAHVDVVQPARLEERFAASTRLMPVRAPGRLPFDHPE
MSMEG_1268|M.smegmatis_MC2_155 DPKRDDR-GWVLSVAHVAVVPLDFVASRFPESTRLVPAHAPGRLVYDHAD
* :: :.: :* :. .*: : :**
Mb1619c|M.bovis_AF2122/97 MVTHARTRLIAKMSYTNIGFALAPKEFALSTLRDIYGAALGYQVDATNLQ
Rv1593c|M.tuberculosis_H37Rv MVTHARTRLIAKMSYTNIGFALAPKEFALSTLRDIYGAALGYQVDATNLQ
MLBr_01224|M.leprae_Br4923 MVTHARARLVAKMSYTNIGFALAPKEFALSTLRDIYGATLGYQVDATNLQ
MAV_3192|M.avium_104 MVDNARARLIAKLSYTNIGFALAPREFALSTLRDIYGAALGYQVDATNLQ
MMAR_2390|M.marinum_M MVTHAHARLVAKMSYTNIGFALTPKEFALSTLRDIYGATLGYQVDTTNLQ
MUL_1567|M.ulcerans_Agy99 MVTHAHARLVAKMSYTNIGFALTPKEFALSTLRDIYGATLGYQVDTTNLQ
MAB_2675|M.abscessus_ATCC_1997 MVAHAHTRLAAKMSYTNIGFALAPQEFTLSTLRDIYCATLGYPVDATNLQ
TH_0810|M.thermoresistible__bu MVEHARSRLVAKLSYTNIGFGLAPEEFALSTLRDIYAAALGHPVDTTNLQ
Mflv_5180|M.gilvum_PYR-GCK IVARALSYLRDRYARSPDPDHLLDDEFTIRELRLAHEAVAGEPLQRDWFR
Mvan_1101|M.vanbaalenii_PYR-1 IVRRAVEYLQSRYADRPDPDRLLGEEFTILQLRRAHEQIAGRPLQRDWFR
MSMEG_1268|M.smegmatis_MC2_155 IIALAVGDVRTRYETAPDPDHLLGDEFTLRDLRLIHEAVTGHALQRDTFR
:: * : : * **:: ** : * :: ::
Mb1619c|M.bovis_AF2122/97 RVLARRRVITQTGTIAQSGRSGGRPAALYRFTDSQLRVTDEFAALRPPGQ
Rv1593c|M.tuberculosis_H37Rv RVLARRRVITQTGTIAQSGRSGGRPAALYRFTDSQLRVTDEFAALRPPGQ
MLBr_01224|M.leprae_Br4923 RVLARRSVIIQTGTVAQSGRSGGRPAALYRFTDSQLRVTDEFAALRPPGN
MAV_3192|M.avium_104 RVLARRGVITATGTIAQSGRSGGRPAALYRFTDAALRVTDEFAALRPPGL
MMAR_2390|M.marinum_M RVLARRKVITQTGTFAQSGRSGGRPAAMYRFTDSQLRVTDEFAALRPPG-
MUL_1567|M.ulcerans_Agy99 RVLARRKVITQTGTFAQSGRSGGRPAAMYRFTDSQLRVTDEFAALRPPG-
MAB_2675|M.abscessus_ATCC_1997 RVLVRRGVLTPTGTTARSGRSGGRPAALYRFTDDRYRVTDEFAALRPPG-
TH_0810|M.thermoresistible__bu RVLERRHVITRTGSTARSGRAGGRPPALYRFTENRYRVTDEYAAFRPPAV
Mflv_5180|M.gilvum_PYR-GCK RAMEP--QLVATGVMTSRGR--GRPAELFRR--P----------------
Mvan_1101|M.vanbaalenii_PYR-1 RTMEP--QLEPTGQLASGAR--GRPAELFHRREPGS--------------
MSMEG_1268|M.smegmatis_MC2_155 RSMES--QLVATGATVSRGR--GRPAELFRRRDTAGRR------------
* : : ** . .* ***. :::
Mb1619c|M.bovis_AF2122/97 L-
Rv1593c|M.tuberculosis_H37Rv L-
MLBr_01224|M.leprae_Br4923 --
MAV_3192|M.avium_104 PG
MMAR_2390|M.marinum_M --
MUL_1567|M.ulcerans_Agy99 --
MAB_2675|M.abscessus_ATCC_1997 --
TH_0810|M.thermoresistible__bu --
Mflv_5180|M.gilvum_PYR-GCK --
Mvan_1101|M.vanbaalenii_PYR-1 --
MSMEG_1268|M.smegmatis_MC2_155 --