For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLGPPATARTRRHTEQEPVDDNATLSAADLLERNRELQSSRAIRLLAATNLSLYATLMERHCADGVVPET ELVVRLERDLDDLGDPQSGLALIKSWAAQGWLHRIVDPRSDQNVCYLTQDARRALDFLRGMRRQDTLATG GSINGIASRLKQVAIRVGNDPARIRKSIEAEIAALHEELDELDNGARPEPDVTDAYDEARAIAMQMERLI TDIGQYGSMIEQATAALDEPIDSNVAYRDRQRQMYADYQAAWDSQGRDSHRAFLRMINDPDQRAEFESDV AAVADALPALDPALRKVMAGFFELVGHQIDEVERIQQRCAQRVKRFTAFGTLEQSRGVARQLNEAIGAAR NLLKSSLIDSRLDLELPLARHAISSVGALSFKIGDLSNPKPAKPAEGDVDLASFAALTTQVDAPAMSDML NSAVARGAVSLPEAVAMVDNAYLGHVIVLWSWALKQPNEASPGSATVRFRSLDGKDREMAVPRLMFTEPI SVGVAQ
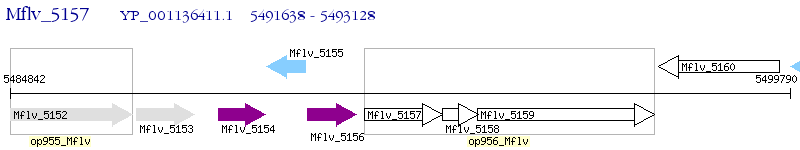
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5157 | - | - | 100% (496) | hypothetical protein Mflv_5157 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1490 | - | 1e-137 | 55.93% (472) | hypothetical protein MAB_1490 |
| M. marinum M | MMAR_3070 | - | 0.0 | 82.84% (472) | hypothetical protein MMAR_3070 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0368 | - | 1e-145 | 56.96% (481) | hypothetical protein MSMEG_0368 |
| M. thermoresistible (build 8) | TH_0008 | - | 0.0 | 77.91% (489) | hypothetical protein MSMEG_0368 |
| M. ulcerans Agy99 | MUL_2315 | - | 1e-161 | 81.95% (349) | hypothetical protein MUL_2315 |
| M. vanbaalenii PYR-1 | Mvan_1174 | - | 0.0 | 88.25% (485) | hypothetical protein Mvan_1174 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5157|M.gilvum_PYR-GCK MLGPPATARTRRHTEQEPVDDNATLSAADLLERNRELQSSRAIRLLAATN
Mvan_1174|M.vanbaalenii_PYR-1 ------------------MHDNSTLSASDLLELNRDLQASRAIRLLATTN
MMAR_3070|M.marinum_M -----------------MHDDNAGLSATELLDLNRDLQGSRAIRLLATTN
MUL_2315|M.ulcerans_Agy99 MLRRPTNYRSR----TQMHGDNAGLSATELLDLNRDLQGSRAIRLLATTN
TH_0008|M.thermoresistible__bu -LDRLIG--AEGDIEDQLVDDNRGMSAADLLGMNRELQGSRAIRLLATHN
MAB_1490|M.abscessus_ATCC_1997 ---------------------MNDLTVEELFAHNDDVQKSRAVRLLAAKN
MSMEG_0368|M.smegmatis_MC2_155 ----------------MPDADVGRPSLQRLHELNREVAESRSVRLLVANN
: * * :: **::***.: *
Mflv_5157|M.gilvum_PYR-GCK LSLYATLMERHCADG-VVPETELVVRLERDLDDLGDP---QSGLALIKSW
Mvan_1174|M.vanbaalenii_PYR-1 LGLYATLMERHLADG-VTPETELVVRLERDLDELDDP---QSGLTLIKSW
MMAR_3070|M.marinum_M LSLYAMLMERHLSDG-ALPETELVVQLERDLEDVNDP---QPGLALIKSW
MUL_2315|M.ulcerans_Agy99 LSLYATLMERHLSDG-ALPETELVVQLDRDLEDVNDP---QPGLALIKSW
TH_0008|M.thermoresistible__bu LGLYATLMERHLNEG-VVAETELVIRLERDLAELDDHPDGQSGLALIKNW
MAB_1490|M.abscessus_ATCC_1997 LAPYITLMERHLDRSAKVSEPQLVAKLDRDLSAVGL--GEQSGLALIKSW
MSMEG_0368|M.smegmatis_MC2_155 LAGYLTLMERHLDRGTKLTEPELVARLERDLADLGAD-GEQSGLALIKYW
*. * ***** . .*.:** :*:*** :. *.**:*** *
Mflv_5157|M.gilvum_PYR-GCK AAQGWLHRIVDPR--SDQNVCYLTQDARRALDFLRGMRRQDTLATGGSIN
Mvan_1174|M.vanbaalenii_PYR-1 ASQGWLHRIVDPR--SDQNVCYLTQDARRALDFLRGMRRQDTIATGGSIN
MMAR_3070|M.marinum_M AAQGWLHRIVDER--HGQNVCYLTQDARRALDFLRGLRRQDTIATGGSIT
MUL_2315|M.ulcerans_Agy99 AAQGWLHRIVDER--HGQNVCYLTQDARRALDFLRGLRRQDTIATGGSIT
TH_0008|M.thermoresistible__bu ASQGWLHRIVDDR--TGQNVCYLTQDARRALDFLRGLRRNDTIATGGSIN
MAB_1490|M.abscessus_ATCC_1997 ASDGWLHRASDGTGPDAENVCSLTEDARSALAFVRRLRRADTVATGGSIA
MSMEG_0368|M.smegmatis_MC2_155 ASQGWLHRITEQAGDGERNVCYLTYEARAVLDFARRMRREDTIATGGSIA
*::***** : .*** ** :** .* * * :** **:******
Mflv_5157|M.gilvum_PYR-GCK GIASRLKQVAIRVGNDPARIRKSIEAEIAALHEELDELDNGAR---PEPD
Mvan_1174|M.vanbaalenii_PYR-1 GIASRLKQVALKVGGDPARIRKSIEAEIEALHRELDELDRGERRTFSETD
MMAR_3070|M.marinum_M GIAARLKQVAVKVDNDPARIRKHIEAEIAALHEELDQLDAGQR---PQTD
MUL_2315|M.ulcerans_Agy99 GIAARLKQVAVKVDNDPARIRKHIEAEIAALHEELDQLDAGQR---PQTD
TH_0008|M.thermoresistible__bu GIAARLKQVAIRVGGDPARIRKDIEAQIAALQAELEALEAGER---PEPD
MAB_1490|M.abscessus_ATCC_1997 GIAAGLKRVATQLDGDPSRLRADIEAQIAELQTQLHAIDDGYR---PEPD
MSMEG_0368|M.smegmatis_MC2_155 GIAAGLRRVAGQVSNDPERIRADIEAEIDRLRAELDALDEGRR---PEPD
***: *::** ::..** *:* ***:* *: :*. :: * * .:.*
Mflv_5157|M.gilvum_PYR-GCK VTDAYDEARAIAMQMERLITDIGQYGSMIEQATA-ALDEPIDSNVAYRDR
Mvan_1174|M.vanbaalenii_PYR-1 ATDAYDEARAIAMQMERLITDIGQYGSMIEQATA-ALDEPIDSNVEYRDR
MMAR_3070|M.marinum_M VTDCYDEARAIALQMERLITDIGQYGTMIERATS-ALDEPIDSNAEYRDR
MUL_2315|M.ulcerans_Agy99 VTDCYDEARAIALQMERLITDIGQYGTMIERATS-ALDEPIDSNAEYRDR
TH_0008|M.thermoresistible__bu VTDCYDEARAIALQMERIITDIGQYGTMIEQATA-ALDEPIDSNAAYRER
MAB_1490|M.abscessus_ATCC_1997 IVDLEDETRAIAYQMEQVITDIVRYGSMQNEITAGLIDAAEDSDSGFRDR
MSMEG_0368|M.smegmatis_MC2_155 PLDVEDEARAIALQMEQIISDIGQYGAMLNRITTRLLDASADSDVGYRER
* **:**** ***::*:** :**:* :. *: :* . **: :*:*
Mflv_5157|M.gilvum_PYR-GCK QRQMYADYQAAWDSQGRDSHRAFLRMINDPDQRAEFESDVAAVADALPAL
Mvan_1174|M.vanbaalenii_PYR-1 QRQMYADYQAAWDSQGRDSHRAFLRMINDPDQRAEFEADVAAVADALPHL
MMAR_3070|M.marinum_M QRQMYSDYQAAWDSQGRDSHRAFLRMINDPDQRAEFEADVAAVAEALPSL
MUL_2315|M.ulcerans_Agy99 QRQMYSDYQAAWDSQGRDSHRAFLRMINDPDQRAEFEADVAAVAEALPRL
TH_0008|M.thermoresistible__bu QRRLYADYQAAWESSGRDSHRAFLRMVNDPDQRAEFEADVAAVAEALPAL
MAB_1490|M.abscessus_ATCC_1997 ARRMFADYDALFDSRERASYSAFTRTIQDPDQRAALRSDIACVAEGLPDL
MSMEG_0368|M.smegmatis_MC2_155 QRQLFDDYESLFASRESASYTAFTRMIQDPEQRAALVADIELVTSQLPQL
*::: **:: : * *: ** * ::**:*** : :*: *:. ** *
Mflv_5157|M.gilvum_PYR-GCK DPALRKVMAGFFELVGHQIDEVERIQQRCAQRVKRFTAFGTLEQSRGVAR
Mvan_1174|M.vanbaalenii_PYR-1 DPALRKVMAGFFELVGHQIDEVERIQQRCAQRVKRFTAFGTLEQSRGVAR
MMAR_3070|M.marinum_M DPALRKVMAGFFELVGHQIDEVERIQQRCAQRVKRFTAFGTMEQSRGVAR
MUL_2315|M.ulcerans_Agy99 DPALCKVMAGFFELVGHQIDEVERIQQRCAQRVKRFTAFGTMEQSRGVAR
TH_0008|M.thermoresistible__bu DPQLRKVMTGFFELVGHQIDEVERIQQRCAQRIKRFTAFGTLEQSRGIAR
MAB_1490|M.abscessus_ATCC_1997 DPGLREVMRNFFKLVSQQIAEVSRIEQRCAQRIRRFFAAGTTEQARGRAR
MSMEG_0368|M.smegmatis_MC2_155 DPSLREVMSGFFTLVSAQTAEVGRTRQRCARRIKRFVASGTLEESRGVTR
** * :** .** **. * ** * .****:*::** * ** *::** :*
Mflv_5157|M.gilvum_PYR-GCK QLNEAIGAARNLLKSSLIDSRLDLELPLARHAISSVGALSFKIGDLSNPK
Mvan_1174|M.vanbaalenii_PYR-1 QLNEAIGAARNLLKSSLIDSRLDIELPLARHTISSVGALSFKIGDLSNPK
MMAR_3070|M.marinum_M QLNEAIAAARALLKSSFTDSRLDIEVPLARHAISSVGALGFRIGDLSNPK
MUL_2315|M.ulcerans_Agy99 QLNEAIAAAAP--CSSRRSPTHAWTSRSRWRGTRSVPSVPSASGSATCPT
TH_0008|M.thermoresistible__bu QLNEAIAAARALLKTSLADSRLDIEVPLTRHAISSVGALSFKISDLSVPK
MAB_1490|M.abscessus_ATCC_1997 QLNDALAAGHALLKMSTADSPIDAELPIGRSAAAAIGALSFTIKDTAPPV
MSMEG_0368|M.smegmatis_MC2_155 QLNDALAAANALLSVSLADSRVGFEVPLARADFNSIGAVAFKIRDAVPPT
***:*:.*. * .. :: :: . *
Mflv_5157|M.gilvum_PYR-GCK PAKPA-EGDVDLASFAALTTQVDAPAMSDMLNSAVARGAVSLPEAVAMVD
Mvan_1174|M.vanbaalenii_PYR-1 PAKPA-GGDVDLTSFAALTTQVDAPPMSDMLNAAVTKGPVSLPEAIEMLG
MMAR_3070|M.marinum_M PAKPG-DTEVDLTSFAAFTTQVDAPAMSDMLNHAITAGPVALPDAVAMLD
MUL_2315|M.ulcerans_Agy99 PSPPN-PATPRSTSPASPP----SPPRSTRP-RCPTCSTTRSPRGLSRYP
TH_0008|M.thermoresistible__bu PAEPA-GGEVDLSSFAALTTQVDATAMAERLNTELASGPVSLPQAVDLVD
MAB_1490|M.abscessus_ATCC_1997 LAQDAPSGPLDLSGFSALATQVDMAALADTVNSAIASGPVSLPDMISHVE
MSMEG_0368|M.smegmatis_MC2_155 PAQPS-EGEVNLSAFGALAAQVDATELAELVNDTVVSRPLPLNEAIGMLD
: :. .: . . : . . :
Mflv_5157|M.gilvum_PYR-GCK N----AYLGHVIVLWS-WALKQPNEA-SPGSATVRFRSLDGKDREMAVPR
Mvan_1174|M.vanbaalenii_PYR-1 RSGQGAYLGHVIVLWS-WALKQPNEH-EAPSTTVRFRSLDGKDREIAVPH
MMAR_3070|M.marinum_M T----AYLGHVIVLWS-WALKQPNPD-TAESTAVRFRSLDGRDLEIAVPR
MUL_2315|M.ulcerans_Agy99 T----PSPCSTPPTWATSSCCGPGPS-NSPTPTP-----PSRPPSASAPW
TH_0008|M.thermoresistible__bu GAGQGAYLGHVIVLWS-WALKQPGVDGDCPSTWVRFRSPDGRDRAIEVPN
MAB_1490|M.abscessus_ATCC_1997 A----PYLGDVIVLWS-LALKQDNTA-DRESVKVKFRSLDGQDRVMEVPQ
MSMEG_0368|M.smegmatis_MC2_155 S----AYLGHVIVLWS-WALKQPAVG-NAESVRVRFRSLEGPDREIEVPK
. . *: : : . .*
Mflv_5157|M.gilvum_PYR-GCK LMFTEPISVGVAQ-----
Mvan_1174|M.vanbaalenii_PYR-1 LMFTEPISVGAAQ-----
MMAR_3070|M.marinum_M LTFTEPINTIAGAST---
MUL_2315|M.ulcerans_Agy99 TAVTSKSRYLV-------
TH_0008|M.thermoresistible__bu LMFTEPITLAGALS----
MAB_1490|M.abscessus_ATCC_1997 LMFREPVLATLEEEGSTL
MSMEG_0368|M.smegmatis_MC2_155 LVFTEPIPIVSE---SLL
. .