For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLAHLSVETGLVHDYGVACATHAADLDAAAARLRGVGVGAAAAFGPVGAPFLASLTRAAAAQSASLGRLS TSVDAGRAAATSCAQAYDLSDGDAASRLAGSI
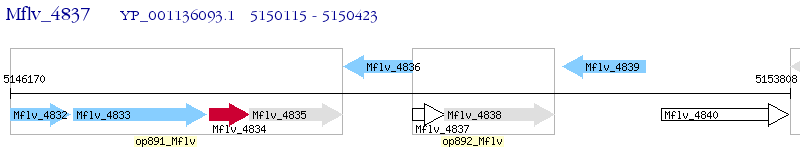
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4837 | - | - | 100% (102) | hypothetical protein Mflv_4837 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3663 | - | 0.0001 | 24.27% (103) | hypothetical protein MAB_3663 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1687 | - | 3e-07 | 36.73% (98) | hypothetical protein MSMEG_1687 |
| M. thermoresistible (build 8) | TH_3491 | - | 4e-06 | 44.26% (61) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4837|M.gilvum_PYR-GCK MLAHLSVETGLVHDYGVACATHAADLDAAAARLRGVGVG-AAAAFGPVGA
MSMEG_1687|M.smegmatis_MC2_155 -MLALFADTGTIRAHGAACAAHTADLAALAAVLRTLPS--HLPSLGPAAD
TH_3491|M.thermoresistible__bu ----MRADTQIIDRFGRAGIRQAAELDAVAAAVSMLPLTAAAPVFGAVGA
MAB_3663|M.abscessus_ATCC_1997 MPEELSADTDLLRGVGGVQMGMGGTLAAVGTAVSIFPVAALAPVFGPIGA
: .:* : * . . * * .: : . . :*. .
Mflv_4837|M.gilvum_PYR-GCK PFLASLTRAAAAQSAS-LG--RLSTSVDAGRAAATSCAQAYDLSD---GD
MSMEG_1687|M.smegmatis_MC2_155 RFAAVFLDALDAQAKA-VA--ALGDQVVQAGVTAQRNAAAYDAAG---HH
TH_3491|M.thermoresistible__bu RFLAALADAAAQQAAXXXGGWRVSPRPHCTARNGSTRVAPLNSSGRTDGR
MAB_3663|M.abscessus_ATCC_1997 HYLGAFAMTSAKQGLD-VG--QLAMNVTESGVTTNVASNGYDDADDNIGK
: . : : *. . :. : :.
Mflv_4837|M.gilvum_PYR-GCK AASRLAGSI--
MSMEG_1687|M.smegmatis_MC2_155 AATLL------
TH_3491|M.thermoresistible__bu PTASAPGRTG-
MAB_3663|M.abscessus_ATCC_1997 NISATVEGIEA
: