For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLDLDDIQHILLTRTPAITGRYEFLTFDRADGGRAWLTELLDRVQSASDAVATMDASQRWITLAFTWTG LRALGVPDDALATFPDEFREGMAARADILGDTGPNAPQNWVDGLAGDDVHAIAILFARDDDEHRRCVGEH DKLVARCDGVRTVSYLDLNASPPFNYAHDHFGFRDRLSQPVIKGSGEEPTPGSGAPLEPGEFILGYPDEF TATPEIPQPDELARNGSYMAYRRLQEHIGAFRSYLADHAGTPDEQELLAAKFMGRWRSGAPLVLSPDVDD PELGADPLRNNNFNYKEMDPFGYACPLGSHARRLNPRDTAHNMNRRRMIRRGATYGPALPDGAPEDGTDR GIAAFIICASLVRQFEFAQNVWINDTTFHELGNEHDPICGTQDGTLDFTVPKRPIRTVHKGIPAFTTLRG GAYFFLPGIKALRYLAGGLR
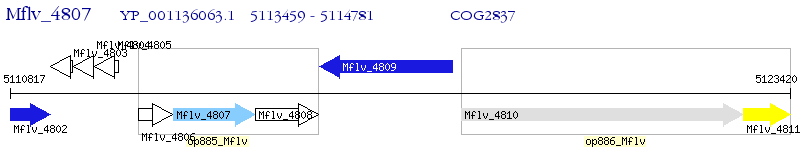
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4807 | - | - | 100% (440) | Dyp-type peroxidase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3700 | - | 0.0 | 79.26% (434) | peroxidase |
| M. thermoresistible (build 8) | TH_2943 | - | 5e-06 | 29.74% (195) | PUTATIVE Dyp-type peroxidase family |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1635 | - | 0.0 | 85.52% (435) | Dyp-type peroxidase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4807|M.gilvum_PYR-GCK MTLDLDDIQHILLTRTPAITGRYEFLTFDRADGGRAWLTELLDRVQSASD
Mvan_1635|M.vanbaalenii_PYR-1 MTLELDDIQHILLTRTPAMTGRYEFLTFDSPAGARAWLTELLGVVHSAAD
MSMEG_3700|M.smegmatis_MC2_155 -MLDLDDIQHILLTRTPAITGRYEFLTFDTPAGGRAWLTALLDKVQSAAD
TH_2943|M.thermoresistible__bu ------------VTGPDGRRGISRRRFVAGTLGAGAAVGAAAMVPGCAGR
:* . . * . . . *. * : .*.
Mflv_4807|M.gilvum_PYR-GCK AVATMDASQRWITLA-FTWTGLRALGVPDDALATFPDEFREGMAARADIL
Mvan_1635|M.vanbaalenii_PYR-1 VTTTMDASKRWVTVA-FTWTGLRALGVPPHALDTFPIEFREGMAARADIL
MSMEG_3700|M.smegmatis_MC2_155 VRASMDTSDRWVTLA-FTWTGLRALGVPEESLSTFPDEFREGMAARASIL
TH_2943|M.thermoresistible__bu QDDSLPDSPRFVPFEGPHQTGITALPVP-----------RLGLLASFKVV
:: * *::.. **: ** ** * *: * .::
Mflv_4807|M.gilvum_PYR-GCK GDTGPNAPQNWVDGLAGDDVHAIAILFARDDDEHRRCVGEHDKLVARCDG
Mvan_1635|M.vanbaalenii_PYR-1 GDTGANAPDHWVGGLAGDDVHAIAVVFARDDEEHARCVGEHDSLVARCDG
MSMEG_3700|M.smegmatis_MC2_155 GDTGANAPEHWPGGLAGTDLHAIAILFSRNEEQCRKSISEHDRLLSRTNG
TH_2943|M.thermoresistible__bu SDDRS----RLALVLRELTDEIRGLMAGRPPEVRDPAYPPVDSGILGAQ-
.* . . * . .:: .* : . * : :
Mflv_4807|M.gilvum_PYR-GCK VRTVSYLDLNASPPFNYAHDHFGFRDRLSQPVIKGSGEEPTPGSGAPLEP
Mvan_1635|M.vanbaalenii_PYR-1 VRTLSYLDLNAHPPFSYAHDHFGFRDRLSQPAIKGSGEEPTPGSGAALEP
MSMEG_3700|M.smegmatis_MC2_155 VRSLSHLDLNASPPFDHAHDHFGFRDRLSQPVMKYSGEEPTPGSGDALEP
TH_2943|M.thermoresistible__bu -PPPDDLSVVVGVGASVFDDRFGLADRRPNELVTMP-FLANDRLDPKLSH
. . *.: . . .*:**: ** .: :. . .. . *.
Mflv_4807|M.gilvum_PYR-GCK GEFILGYPDEFTATPEIPQPDELARNGSYMAYRRLQEHIGAFRSYLADHA
Mvan_1635|M.vanbaalenii_PYR-1 GEFILGYPDEAGPVADLPEPEDLSRNGSYMAYRRLQEHVGTFRSYLADNA
MSMEG_3700|M.smegmatis_MC2_155 GEFILGYPDESGPVAHLPQPVVLSRNGSFMAYRRLQEHVGVFREYLREHS
TH_2943|M.thermoresistible__bu GDLLLSLEAGHTDTLQFALRQLMRRTRRDLVLHWMID--GYARGAGSGVP
*:::*. . .:. : *. :. : : : * * .
Mflv_4807|M.gilvum_PYR-GCK GTPDEQELLAAKFMGRWRSGAPLVLSPDVDDPELGADPLRNNNFNYKEMD
Mvan_1635|M.vanbaalenii_PYR-1 GTPEEQELLAAKFMGRWRSGAPLVLCPDADDPELGADPQRNNDFNYKEMD
MSMEG_3700|M.smegmatis_MC2_155 DDPAGEALLAAKFMGRWRSGAPLVLAPECDDPALGADPMRNNNFNYKDMD
TH_2943|M.thermoresistible__bu TETPRNLLGFKDGTANLDSGDAALMDRYVWVGSDDGEPDWAVGGSYQVVR
. : * . .. ** . :: ..:* . .*: :
Mflv_4807|M.gilvum_PYR-GCK PFGYACPLGSHARRLNPRDTAHNMNRRRMIRRGATYGPALPDGAPEDGTD
Mvan_1635|M.vanbaalenii_PYR-1 PFGYACPLGSHARRLNPRDTAHYMNRRRMIRRGATYGPALPDGAPDDGVD
MSMEG_3700|M.smegmatis_MC2_155 PFGYACPLGSHARRLNPRDTAHYMNRRRMIRRGATYGPALPEGAPDDGVE
TH_2943|M.thermoresistible__bu LIRMFVEFWDRTR-LAEQEAIFGRHKVSGAPLGMSDEFDDPDYASDPDGE
: : .::* * ::: . :: * : *: *.: . :
Mflv_4807|M.gilvum_PYR-GCK RGIAAFIICASLVRQFEFAQNVWINDTTFHELGNEHDPICGTQDGTLDFT
Mvan_1635|M.vanbaalenii_PYR-1 RGIAAFIICASLVRQFEFAQNVWINDQTFHELGNEHDPICGSQDGTFDFT
MSMEG_3700|M.smegmatis_MC2_155 RGIAAFIICADLVRQFEFAQNVWINDKTFHELGNEHDPICGTQDGTMDFT
TH_2943|M.thermoresistible__bu RIPLNAHIRLANPRTPETEDNLLLRRGFSYARGFDG---AGRLDQGLAFV
* * * * :*: :. : * : .* * : *.
Mflv_4807|M.gilvum_PYR-GCK VPKRPIRT----VHKGIP------AFTTLRGGAYFFLPGIK-ALRYLAGG
Mvan_1635|M.vanbaalenii_PYR-1 VPKRPIRR----VHKGIP------AFTTLRGGAYFFLPGLT-ALRYLTTL
MSMEG_3700|M.smegmatis_MC2_155 VPKRPIRK----VHKGLP------AFTSVRGGAYFFLPGIS-ALRYLAAL
TH_2943|M.thermoresistible__bu CYQRSLRRGFLAVQERLKGEPLEEYILPVGGGFFFALPGVSEPDRYLGDA
:*.:* *:: : : .: ** :* ***:. . ***
Mflv_4807|M.gilvum_PYR-GCK LR----
Mvan_1635|M.vanbaalenii_PYR-1 DD----
MSMEG_3700|M.smegmatis_MC2_155 KDEVTA
TH_2943|M.thermoresistible__bu LVD---