For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTEKNSGPQEGIKGVVEDVKGKAKETIGTVAGRDDMVQEGKAQQDKADAQREVAEKEAEAESARAGAKAA EKRQEANQ
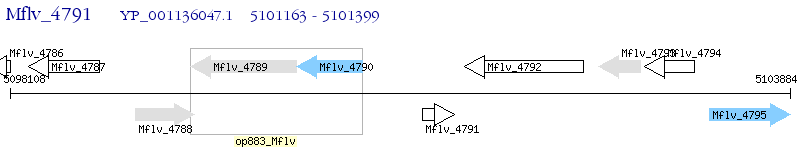
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4791 | - | - | 100% (78) | hypothetical protein Mflv_4791 |
| M. gilvum PYR-GCK | Mflv_2022 | - | 1e-26 | 74.32% (74) | CsbD family protein |
| M. gilvum PYR-GCK | Mflv_3881 | - | 1e-21 | 62.50% (72) | CsbD family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2518 | - | 1e-29 | 74.36% (78) | hypothetical protein MAB_2518 |
| M. marinum M | MMAR_2989 | - | 9e-25 | 69.86% (73) | hypothetical protein MMAR_2989 |
| M. avium 104 | MAV_3734 | - | 7e-28 | 72.37% (76) | hypothetical protein MAV_3734 |
| M. smegmatis MC2 155 | MSMEG_1770 | - | 8e-34 | 87.18% (78) | hypothetical protein MSMEG_1770 |
| M. thermoresistible (build 8) | TH_3361 | - | 1e-29 | 78.38% (74) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_2228 | - | 5e-24 | 68.49% (73) | hypothetical protein MUL_2228 |
| M. vanbaalenii PYR-1 | Mvan_1657 | - | 7e-35 | 87.18% (78) | CsbD family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1770|M.smegmatis_MC2_155 --------------------MTEK-NSGPEEGIKGAVEDVKGKAKEAIGT
Mvan_1657|M.vanbaalenii_PYR-1 MGWLTEGAPKPHSPIERKAIMTEK-NSGPEEGIKGIVEDVKGKAKETVGT
Mflv_4791|M.gilvum_PYR-GCK --------------------MTEK-NSGPQEGIKGVVEDVKGKAKETIGT
MAB_2518|M.abscessus_ATCC_1997 --------------------MTEK-NSGPQEAVRGVVEDVKGKAKEAAGT
TH_3361|M.thermoresistible__bu --------------------MAD---TGPQEGIKGVVEDAKGKTKEAVGT
MMAR_2989|M.marinum_M --------------------MAD--KSGPQEAVEGVVEGAKGKAKEVFGA
MUL_2228|M.ulcerans_Agy99 --------------------MAD--KSGPQEAVEGVVEGAKGKAKEVFEA
MAV_3734|M.avium_104 --------------------MAEENKSGPAEAVKGVVEDVKGKAKEAVGA
*:: :** *.:.* **..***:**. :
MSMEG_1770|M.smegmatis_MC2_155 VAGRDDMVREGKAQQDKADAQQAAAKKEAEAESARAGAKAAEKRQESEQR
Mvan_1657|M.vanbaalenii_PYR-1 VTGRDDMVREGKAQQDKADAQQDAAKKEAEAESARAGAAAAEKRQEANQ-
Mflv_4791|M.gilvum_PYR-GCK VAGRDDMVQEGKAQQDKADAQREVAEKEAEAESARAGAKAAEKRQEANQ-
MAB_2518|M.abscessus_ATCC_1997 VTGRDDLIEEGKAQQDKAQAQREAAQKEAEAEKSRAEAKSDEKRQKAHE-
TH_3361|M.thermoresistible__bu VTGRDDMINEGKAQQDKADAQRDAAKKEAEAESARGGAKAAEERQKENQ-
MMAR_2989|M.marinum_M VTGRDDVKREGQAQQDKADAQRDAAKKEAEAEAARRGADVAEERQKANQ-
MUL_2228|M.ulcerans_Agy99 VTGRDDVKREGQAQQDKADAQRDAAKKEAEAEAARRGADVAEERQKANQ-
MAV_3734|M.avium_104 VAGRDDLTREGQAQQDKAEAQRDAAKKEAEAEAARGGAEAAEERQRANQ-
*:****: .**:******:**: .*:****** :* * *:**. .:
MSMEG_1770|M.smegmatis_MC2_155 P
Mvan_1657|M.vanbaalenii_PYR-1 -
Mflv_4791|M.gilvum_PYR-GCK -
MAB_2518|M.abscessus_ATCC_1997 -
TH_3361|M.thermoresistible__bu -
MMAR_2989|M.marinum_M -
MUL_2228|M.ulcerans_Agy99 -
MAV_3734|M.avium_104 -