For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ERKGFKKIAAGTASFGMLLTGVLVAAPTAGAQTSTWTMPALRGEVLERAVNSVEGAAGSAPIRFNIYDSV NNQVVYNYTNWVVCGQSPRAEATVKVGAKPQTVTFALKRRQAGC*
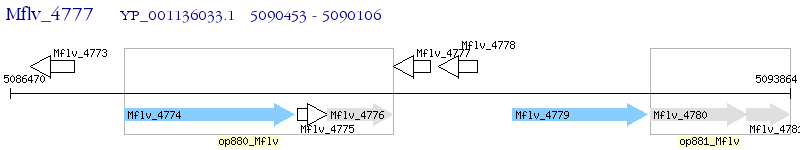
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4777 | - | - | 100% (115) | hypothetical protein Mflv_4777 |
| M. gilvum PYR-GCK | Mflv_4778 | - | 2e-12 | 36.84% (114) | hypothetical protein Mflv_4778 |
| M. gilvum PYR-GCK | Mflv_3070 | - | 6e-05 | 34.34% (99) | hypothetical protein Mflv_3070 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1762 | - | 9e-07 | 29.91% (107) | hypothetical protein MSMEG_1237 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1672 | - | 5e-35 | 57.38% (122) | hypothetical protein Mvan_1672 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4777|M.gilvum_PYR-GCK MERKGFKK----IAAGTASFGMLLTGVLVAAPTAGAQTS---TWTMPALR
Mvan_1672|M.vanbaalenii_PYR-1 MELKGFKRQQRNLAVAAAAAGVFAAGIGISAPIAAAAPTAGETWTMGSYR
TH_1762|M.thermoresistible__bu MNRP--------SVTGVASLGLLVAGAIALAPAAAADET----WTMPNLI
*: ....*: *:: :* ** *.* : ***
Mflv_4777|M.gilvum_PYR-GCK GEVLERAVNSVEGAAGSAPIRFNIYDSVNN--QVVYNYTNWVVCGQSPRA
Mvan_1672|M.vanbaalenii_PYR-1 GEVLQRAIDDVIANAGEQNVRFNLYDRVLN--QVIYNYTNWVVCGQSPRA
TH_1762|M.thermoresistible__bu GTDLQGAQDAIQSLTDG-AVWFSSSTDLTGKGRLQIIDRNWTVCSSTPPP
* *: * : : . :. : *. : . :: **.**..:* .
Mflv_4777|M.gilvum_PYR-GCK EATVKVGAKPQTVTFALKRRQAGC-
Mvan_1672|M.vanbaalenii_PYR-1 DASVKVGEKPQTVTFALSRRSTGC-
TH_1762|M.thermoresistible__bu GATFTASTR---IDFGVVRDTEDCP
*:.... : : *.: * .*