For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RDTRIPAVSRFGITGRAAAAYLLLAGLQRGVSLLILPFISHAMLPSEYGAASTLTASAMLIVAIIAAPVE QLVFRAAARGGDDAPALIRVAGTYCYLVLPLFIACVAAGSVVYLPDLFGISGQIWAIELLAVGFLPAATY FALPFVRARQDLRKFVWLAATSILVTALSKFLLIVVWGLGVFGWAISDLVSAVVSALLAIALIRLPRVHV AARNICAVLTFSIPLIPHRASFWALSSLSRPAMATVSTLAQVGLLSFGLNLAAVANMILAELNQAFQPRY SREKFPAPNGETRKPVHWQLVLAFAIPALVGASVALGGQFIFAESYWPSFALTGLLLCGQVAYGLYLVPM NYIVQSAGLPKFSALASVSGATIILVGILALGRQYGAVGVAYVTSSGFAAMAVVAFILTRILRLNISWSS WLKNWPEIALGLAGLVCSIAALVLIVGSTASYIFAVASIVLTVYALALTAARRSS*
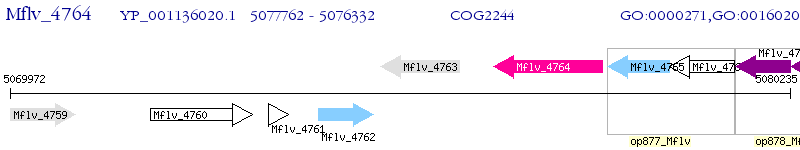
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4764 | - | - | 100% (476) | polysaccharide biosynthesis protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4675 | - | 5e-05 | 22.77% (448) | hypothetical protein MUL_4675 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4764|M.gilvum_PYR-GCK --MRDTRIPAVSRFGITGRAAAAYLLLAG--LQRGVSLLILPFISHAMLP
MUL_4675|M.ulcerans_Agy99 MTEPTAPSPVVPSVPVARLAQAFSIQLICRILGMLASVVSVMLTARYLGP
: *.*. . :: * * : * * .*:: : : :: : *
Mflv_4764|M.gilvum_PYR-GCK SEYGAASTLTASAMLIVAIIAAPVEQLVFRAAARGGDDAPALIRVAG-TY
MUL_4675|M.ulcerans_Agy99 GRYGQLTVAVAFIGMWASLVDLGIGRVIVRRVTSSPDDLERLVRIYNGLS
..** :. .* : .::: : :::.* .: . ** *:*: .
Mflv_4764|M.gilvum_PYR-GCK CYLVLPLFIACVAAGSVVYLPDLFGISGQIWAIELLAVGFLP-----AAT
MUL_4675|M.ulcerans_Agy99 LVYCIPLTLVAAGSGLLVYHDHEVRAMLVVLSCQLLIVAMLTRLEPVFLV
:** :....:* :** . . : : :** *.:*. .
Mflv_4764|M.gilvum_PYR-GCK YFALPFVRARQDLRKFVWLAATSILVTALSKFLLIVVWGLGVFGWAISDL
MUL_4675|M.ulcerans_Agy99 TVRFTAVAVSDVVGRLATLGVISFLVASRPDVIWFAVPQLIQTALQLLIQ
. :. * . : : ::. *.. *:**:: ...: :.* * . :
Mflv_4764|M.gilvum_PYR-GCK VSAVVSALLAIALIRLPRVHVAARNICAVLTFSIPLIPHRASFWALSSLS
MUL_4675|M.ulcerans_Agy99 SRAASQRVRLRPILALR----EAADLLRESLFPMGVTVITVLYWRVDGVI
*. . : .:: * * :: *.: : . :* :..:
Mflv_4764|M.gilvum_PYR-GCK RPAMATVSTLAQVGLLS-FGLNLAAVANMILAELNQAFQPRYSREKFPAP
MUL_4675|M.ulcerans_Agy99 LSLLNTHSEVGVYGLASSIALNTLVLSVFFLKSTLSTATELYSR-DLAAF
. : * * :. ** * :.** .:: ::* . .: *** .:.*
Mflv_4764|M.gilvum_PYR-GCK NGETRKPVHWQLVLAFAIPALVGASVALGGQFIFAESYWPSFALTGLLLC
MUL_4675|M.ulcerans_Agy99 AGFIRRSVELMYFLVVPIAVVGVLVAQPLIRLLSSQAFVSRGTPTLALLF
* *:.*. .*...*..: . ::: :::: . : * **
Mflv_4764|M.gilvum_PYR-GCK GQVAYGLYLVPMNYIVQSAGLPKFSALASVSGATIILVGILALGRQYGAV
MUL_4675|M.ulcerans_Agy99 IAVGLRFIATTLGEGLFASHSQRFWFWLSVATLTLNIVLNLALDGRLGAV
*. : ..:. : :: :* **: *: :* ***. : ***
Mflv_4764|M.gilvum_PYR-GCK GV-AYVTSSGFAAMAVVAFILTRILRLNISWSSWLKNWPEIALGLAGLVC
MUL_4675|M.ulcerans_Agy99 GAGIALVCTELFNMTIASWWLHRQCGYHTPVLFLLRLSVPTAASVAVVLP
*. :..: : *::.:: * * : . *: * .:* ::
Mflv_4764|M.gilvum_PYR-GCK SIAALVLIVGSTASYIFAVASIVLTVYALALTAARRSS--------
MUL_4675|M.ulcerans_Agy99 LAGHHVIVVLAAAAVAYLAINAAIGPLTWSTMASLRPNAQPEQLTP
. *::* ::*: : . . .: : : *: *..