For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GFMCGRFAVTTDPALLAEKIKAIDESTAASKDRPTADYPTANYPTANYNVAPTTAVATVVKRHSEPEDES TRRVRLMRWGLVPPWVKAGEDGGPDTKSGPLLINARSDKVTSSPAFRSSAKTKRCLVPMDGWYEWKGTKG AKTPYYMYAGDGEPLFMAGLWTTWRPKPADTDAPKDAKPLLSCTIITTDAAAQLADIHDRMPLTISEPDW DRWLDPDAPIDEGLLRGHGDLDRIEIREVSRLVNSVRNNGPELIEPVTPQAEQGTLL*
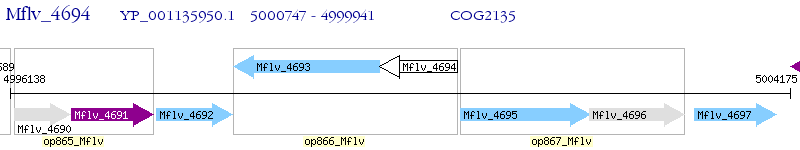
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4694 | - | - | 100% (268) | hypothetical protein Mflv_4694 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3255c | - | 1e-86 | 58.46% (272) | hypothetical protein Mb3255c |
| M. tuberculosis H37Rv | Rv3226c | - | 1e-86 | 58.46% (272) | hypothetical protein Rv3226c |
| M. leprae Br4923 | MLBr_00793 | - | 2e-85 | 58.55% (275) | putative bacteriophage protein |
| M. abscessus ATCC 19977 | MAB_3545c | - | 2e-78 | 54.48% (268) | hypothetical protein MAB_3545c |
| M. marinum M | MMAR_1321 | - | 9e-89 | 59.71% (278) | hypothetical protein MMAR_1321 |
| M. avium 104 | MAV_4181 | - | 1e-100 | 66.54% (272) | hypothetical protein MAV_4181 |
| M. smegmatis MC2 155 | MSMEG_1891 | - | 1e-113 | 72.26% (274) | hypothetical protein MSMEG_1891 |
| M. thermoresistible (build 8) | TH_0662 | - | 1e-115 | 72.63% (274) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2559 | - | 3e-87 | 59.64% (275) | hypothetical protein MUL_2559 |
| M. vanbaalenii PYR-1 | Mvan_1772 | - | 1e-130 | 83.58% (268) | hypothetical protein Mvan_1772 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4694|M.gilvum_PYR-GCK MGFMCGRFAVTTDPALLAEKIKAIDESTAASK------DRPTADYPTANY
Mvan_1772|M.vanbaalenii_PYR-1 MDTMCGRFAVTTDPALLAEKIKAIDETAAAKT------DGP---------
MSMEG_1891|M.smegmatis_MC2_155 MGLMCGRFAVTTDPALLAQKIQAIDETAAASSG-----DKDVAG------
TH_0662|M.thermoresistible__bu MGYMCGRFAVTTDPALLAQKIQAIDESRAAADAQGGNPQSPGRGSAAPNY
MMAR_1321|M.marinum_M MGVMCGRFAVTTDPALLAEKINAIDEASGAASD----------------G
MUL_2559|M.ulcerans_Agy99 ---MCGRFAVITDPALLAEKINAIDEASGAASD----------------G
Mb3255c|M.bovis_AF2122/97 ---MCGRFAVTTDPAQLAEKITAIDEATGCG------------------G
Rv3226c|M.tuberculosis_H37Rv ---MCGRFAVTTDPAQLAEKITAIDEATGCG------------------G
MLBr_00793|M.leprae_Br4923 MAGVCGRFAVITSTALLAEKIKAVDEATVDA-------------------
MAV_4181|M.avium_104 ---MCGRFAVTTDPAQLAQKIKAIDETTGAAPS----------------D
MAB_3545c|M.abscessus_ATCC_199 ---MCGRYAVTIDPALLAAEIDAVDETATSAVQS---------------P
:***:** ..* ** :* *:**:
Mflv_4694|M.gilvum_PYR-GCK PTANYNVAPTTAVATVVKRHSEPEDESTRRVRLMRWGLVPPWVKAGEDGG
Mvan_1772|M.vanbaalenii_PYR-1 ---NYNVAPTTSVATVVKRHTEPDDESTRRIRLMRWGLVPPWAKAGDDGS
MSMEG_1891|M.smegmatis_MC2_155 --ANYNVAPTTTITSVVKRHTEPEDESTRRLRSMRWGLIPPWVKATEDGS
TH_0662|M.thermoresistible__bu AAPNYNVAPTTDVATVVRRHHEPDDQPTRRVRLMRWGLIPPWAKTAEDGR
MMAR_1321|M.marinum_M FAPNYNVAPTDMIATVVTRHSEPDDEPTRRVRMMRWGFVPSWAKPGPQGA
MUL_2559|M.ulcerans_Agy99 FAPNYNVAPTDMIATVVTRHSEPDDEPTRRVRMMRWGFVPSWAKPGPQGA
Mb3255c|M.bovis_AF2122/97 GKTSYNVAPTDTIATVVSRHSEPDDEPTRRVRLMRWGLIPSWIKAGPGGA
Rv3226c|M.tuberculosis_H37Rv GKTSYNVAPTDTIATVVSRHSEPDDEPTRRVRLMRWGLIPSWIKAGPGGA
MLBr_00793|M.leprae_Br4923 --PNYNVAPTVPIATVVSRHSESDYEPTRRVRLMRWGLIPPWIKASSDGG
MAV_4181|M.avium_104 TAPNYNVAPTSTIATVVSRHSEPEDEPTRRVRLMRWGLVPPWAKAGPGGA
MAB_3545c|M.abscessus_ATCC_199 TIPNYNVAPTDPVLAVVARHHQPGDVPTRRIRAMRWGLVPNWATARQDGR
.****** : :** ** :. .***:* ****::* * .. *
Mflv_4694|M.gilvum_PYR-GCK PDTKSGPLLINARSDKVTSSPAFRSSAKTKRCLVPMDGWYEWK-------
Mvan_1772|M.vanbaalenii_PYR-1 PDTKSGPLLINARSDKVTSSPAFRSSAKAKRCLVPMDGWYEWK-------
MSMEG_1891|M.smegmatis_MC2_155 PENK-GPLLINARAEKVTTSPAFRSSAKSRRCLIPMDGWYEWRPNP-AAD
TH_0662|M.thermoresistible__bu PDTK-GPLLINARADRVTTSPAFRNSAKSRRCLVPMDGWYEWR-------
MMAR_1321|M.marinum_M PEGK-GPLLINARAETVATSPAFRGAAQHKRCLVPMDGWYEWRANPDVLS
MUL_2559|M.ulcerans_Agy99 PEGK-GPLLINARAETVATSPAFRGAAQHKRCLVPMDGWYEWRANPDVLS
Mb3255c|M.bovis_AF2122/97 PDAK-GPPLINARADKVATSPAFRSAVRSKRCLVPMDGWYEWRVDPDATP
Rv3226c|M.tuberculosis_H37Rv PDAK-GPPLINARADKVATSPAFRSAVRSKRCLVPMDGWYEWRVDPDATP
MLBr_00793|M.leprae_Br4923 PDIK-GPLLINVRADKVATSPVFRNSAKSKRCLVPMDGWYEWQVNVDSAL
MAV_4181|M.avium_104 PETK-GPMLINARADKVTSSPAFRASAKSKRCLIPMDGYYEWRVNSDPAA
MAB_3545c|M.abscessus_ATCC_199 PESK-GAPLINARAETLTTAATFRAAAQSKRCLVPMDGWYEWR-------
*: * *. ***.*:: ::::..** :.: :***:****:***:
Mflv_4694|M.gilvum_PYR-GCK GTKG---AKTPYYMYAGDGEPLFMAGLWTTWRPKPADTDAPKDAKPLLSC
Mvan_1772|M.vanbaalenii_PYR-1 GQKG---AKTPYYMHTRDGEPLFMAGLWSTWRPKGAD----KDVKPLLSC
MSMEG_1891|M.smegmatis_MC2_155 GKKA---SKTPFFMYGVDGEPLFMAGLWTTWRPPGAP----KDHAPLVSC
TH_0662|M.thermoresistible__bu GEKG---AKTPFYLHSIDGEPLFMAGLWSTWRPNGAAG----STTPLLSC
MMAR_1321|M.marinum_M GAGSKKVAKTPFFIHRADGNTVCMAGLWSVWKPNNAA-------APLLSA
MUL_2559|M.ulcerans_Agy99 GAGSKKVAKTPFFIHRADGNTVCMAGLWPVWKPNNAA-------APLLSA
Mb3255c|M.bovis_AF2122/97 GRPN---AKTPFFLHRHDGALLFTAGLWSVWKSYRSA-------PPLLSC
Rv3226c|M.tuberculosis_H37Rv GRPN---AKTPFFLHRHDGALLFTAGLWSVWKSYRSA-------PPLLSC
MLBr_00793|M.leprae_Br4923 GKKA---STTPFFINRDDGATLFMAGLWSVWQPANSS-------AMLLSC
MAV_4181|M.avium_104 GKKP---RKTPFFMYSEDGEPLFMAGLWSVWKPAKDA-------PPLLSC
MAB_3545c|M.abscessus_ATCC_199 KQEG---AKTAFYMNAGEGKRLFAAGLWSVWKPAKGA-------VPLLSC
.*.::: :* : ****..*:. *:*.
Mflv_4694|M.gilvum_PYR-GCK TIITTDAAAQLADIHDRMPLTISEPDWDRWLDPDAPIDEGLLRGHGDLDR
Mvan_1772|M.vanbaalenii_PYR-1 TIITTDAVGPLADIHDRMPLTISAPDWDRWLDPDAPVDEGLLRGHGDLDR
MSMEG_1891|M.smegmatis_MC2_155 TIITTDAAGPLAEIHDRMPLSISRDDWDRWLDPDAPVDEGLLRGHGDLDR
TH_0662|M.thermoresistible__bu AIITTDAVGPLAEIHDRMPLSISQDDWDRWLDPDAPIDEGLLRGHGDLDR
MMAR_1321|M.marinum_M TIITTDAAGELAGIHDRMPLMLSEGDWDAWLNPDAPLDPALLSHPPDVRD
MUL_2559|M.ulcerans_Agy99 TIITTDAAGELAGIHDRMPLMLSEGDWDAWLNPDAPLDPALLSHPPDVRD
Mb3255c|M.bovis_AF2122/97 TVITTDAVGELAEIHDRMPLLLAEEDWDDWLNPDAPPDPELLARPPDVRD
Rv3226c|M.tuberculosis_H37Rv TVITTDAVGELAEIHDRMPLLLAEEDWDDWLNPDAPPDPELLARPPDVRD
MLBr_00793|M.leprae_Br4923 TIITTDAVGELAEIHDRMPLILAEYDWDVWLNPYIPLDPQLLARSPDVHD
MAV_4181|M.avium_104 TIITTDAPGELAQIHDRMPLVMPERDWDRWLDPDAPVDEELLTRPPDVHA
MAB_3545c|M.abscessus_ATCC_199 TIVTTDAVGPLREIHDRMPLMLGADSWDSWLDPDRELDPGLLRAPGSAAG
:::**** . * ******* : .** **:* * ** .
Mflv_4694|M.gilvum_PYR-GCK IEIREVSRLVNSVRNNGPELIEP---VTPQAEQGTLL
Mvan_1772|M.vanbaalenii_PYR-1 IEVREVSRLVNSVRNNGPELIEP---VTPQAEQGTLL
MSMEG_1891|M.smegmatis_MC2_155 IGVREVSRLVNSIRNNGPELIEP---AEPEAEQAGLF
TH_0662|M.thermoresistible__bu IAIREVSKLVNNVRNNGPELIEP---VDPQPEQATLL
MMAR_1321|M.marinum_M MAFREVSTLVNSVRNNGPELLEP---AKPQPEQITLL
MUL_2559|M.ulcerans_Agy99 IAFREVSTLVNSVRNNGPELLEP---AKPQPEQITLL
Mb3255c|M.bovis_AF2122/97 IALRQVSTLVNNVRNNGPELLEP---ARSQPEQIQLL
Rv3226c|M.tuberculosis_H37Rv IALRQVSTLVNNVRNNGPELLEP---ARSQPEQIQLL
MLBr_00793|M.leprae_Br4923 IDVRPVSTLVNSVRNNGPELLEP---VLLQPGQITLL
MAV_4181|M.avium_104 IRMREVSTLVNNVRNNGPELLEP---AKPEPEQARLL
MAB_3545c|M.abscessus_ATCC_199 IETRRVSSLVNSVANNGPELLVPDDGAAAVGEQITLL
: * ** ***.: ******: * . * *: