For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MILFWVMAVVSVGGALGVIAAPKAVYSAMSLAATMIALAVLYIAQGAPFLGVVQIVVYTGAVMMLFLFVL MMIGVDSSESLVETLRGQRIAALAVGLGFGILLIAGIGSVSATGIVGLDQANGGGNVEGLAALIFTKYLW AFELTGALLITAALGAMVLAHRERFQRRKTQRELAVERFAPGGHPTTLPNPGVYARHNAVDVPARLPDGS GSELSVSAILQVRPVPTDERSPGGEE
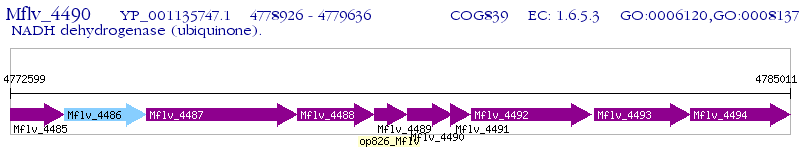
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4490 | - | - | 100% (236) | NADH dehydrogenase subunit J |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3178 | nuoJ | 2e-88 | 72.97% (222) | NADH dehydrogenase subunit J |
| M. tuberculosis H37Rv | Rv3154 | nuoJ | 2e-88 | 72.97% (222) | NADH dehydrogenase subunit J |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2143 | - | 3e-84 | 70.04% (227) | NADH dehydrogenase subunit J |
| M. marinum M | MMAR_1474 | nuoJ | 2e-90 | 71.01% (238) | NADH dehydrogenase I (chain J) NuoJ (NADH- ubiquinone |
| M. avium 104 | MAV_4042 | - | 1e-89 | 70.66% (242) | NADH dehydrogenase subunit J |
| M. smegmatis MC2 155 | MSMEG_2054 | - | 3e-95 | 78.57% (224) | NADH dehydrogenase subunit J |
| M. thermoresistible (build 8) | TH_1088 | nuoJ | 1e-92 | 75.34% (223) | PROBABLE NADH DEHYDROGENASE I (CHAIN J) NUOJ |
| M. ulcerans Agy99 | MUL_2468 | nuoJ | 1e-90 | 71.01% (238) | NADH dehydrogenase subunit J |
| M. vanbaalenii PYR-1 | Mvan_1876 | - | 1e-114 | 88.51% (235) | NADH dehydrogenase subunit J |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3178|M.bovis_AF2122/97 --------MTAVLAS----DVIVRTSTGEAVMFWVLSALALLGAVGVVLA
Rv3154|M.tuberculosis_H37Rv --------MTAVLAS----DVIVRTSTGEAVMFWVLSALALLGAVGVVLA
MMAR_1474|M.marinum_M --------MTSTLASSLAADTITVTSTGEAVTFWILGALALVGALGVVMA
MUL_2468|M.ulcerans_Agy99 --------MTSTLASSLAADTITVTSTGEAVTFWILGALALVGALGVVMA
MAV_4042|M.avium_104 MSAFAGHAVAATLASSYSQDTIVRTSTGEAVTFWVLGALAVIGALGVVLA
Mflv_4490|M.gilvum_PYR-GCK -----------------------------MILFWVMAVVSVGGALGVIAA
Mvan_1876|M.vanbaalenii_PYR-1 --------------------------MGEAVLFWVFAVVSVTGALGVVAA
MSMEG_2054|M.smegmatis_MC2_155 ---MNSDLMLLAAEG-------ARTSTSEAVVFWVVGTVALVGAIGVVAA
TH_1088|M.thermoresistible__bu ---VIGAVITFAAAANDDAVNTVHTSTGEAVLFWIVGGLAVVGALGVVTA
MAB_2143|M.abscessus_ATCC_1997 ----MSLGFWASHATVLAAEGLSRTPTWEGVTFWVLGAIAVLGALGVIAA
: **:.. ::: **:**: *
Mb3178|M.bovis_AF2122/97 VNAVYSAMFLAMTMIILAVFYMAQDALFLGVVQVVVYTGAVMMLFLFVLM
Rv3154|M.tuberculosis_H37Rv VNAVYSAMFLAMTMIILAVFYMAQDALFLGVVQVVVYTGAVMMLFLFVLM
MMAR_1474|M.marinum_M VNAVYSAMFLAMTMIILAVFYMIQDALFLGVVQVVVYTGAVMMLFLFVLM
MUL_2468|M.ulcerans_Agy99 VNAVYSAMFLAMTMIILAVFYMIQDALFLGVVQVVVYTGAVMMLFLFVLM
MAV_4042|M.avium_104 VNAVYSAMFLAMTMIILAVFYMIQDALFLGVVQVVVYTGAVMMLFLFVLM
Mflv_4490|M.gilvum_PYR-GCK PKAVYSAMSLAATMIALAVLYIAQGAPFLGVVQIVVYTGAVMMLFLFVLM
Mvan_1876|M.vanbaalenii_PYR-1 PKAVYSAISLATTMIALAVLYIAQDAPFLGVVQIVVYTGAVMMLFLFVLM
MSMEG_2054|M.smegmatis_MC2_155 RKAVYSAVFLACTMISLAVLYIAQDAPFLGVVQIVVYTGAVMMLFLFVLM
TH_1088|M.thermoresistible__bu PKAVYSALFLAMTMIGLAVLYLAQDAPFLGVVQVVVYTGAVMMLFVFVMM
MAB_2143|M.abscessus_ATCC_1997 PKAVYSAIFLAMTMVILAVFFVAQGALFLGVAQVVVYTGAVMMLFLFVLM
:*****: ** **: ***::: *.* ****.*:***********:**:*
Mb3178|M.bovis_AF2122/97 LIGVDSAESLKETLRGQRVAAVLTGVGFGVLLISTIGQVATR-GFAGLTV
Rv3154|M.tuberculosis_H37Rv LIGVDSAESLKETLRGQRVAAVLTGVGFGVLLISTIGQVATR-GFAGLTV
MMAR_1474|M.marinum_M LIGVDSAESLKETLRGQRVAALVIGVGFGVLLIAAIGNVAAG-GFAGLTA
MUL_2468|M.ulcerans_Agy99 LIGVDSAESLKETLRGQRVAALVIGVGFGVLLIAAIGNVAAG-GFAGLTA
MAV_4042|M.avium_104 LIGVDSAESLKETLRGQRVAAVAAGVGFGILLIAAVGNVASG-GFAGLTT
Mflv_4490|M.gilvum_PYR-GCK MIGVDSSESLVETLRGQRIAALAVGLGFGILLIAGIGSVSAT-GIVGLDQ
Mvan_1876|M.vanbaalenii_PYR-1 LIGVDSSESLVETLRGQRVAAIVVGLGFGVLLVAGVGNVSAT-GFVGLDQ
MSMEG_2054|M.smegmatis_MC2_155 LIGVDLTESLVETIKGHRIAALIAGAGFGILVIAGIGNVSVA-GFSGLAA
TH_1088|M.thermoresistible__bu LIGVDSSEALVETIRGQRVAAVAAGFGFGILLIAGIGSVAAEIGFTGLDE
MAB_2143|M.abscessus_ATCC_1997 FVGVDSSDSLVETLPGQRVGAVAAGLGFGILAIASIGNASTT-VFVGLGQ
::*** :::* **: *:*:.*: * ***:* :: :*..: : **
Mb3178|M.bovis_AF2122/97 ANANGNVEGLAALIFSRYLWAFELTSALLITAAVGAMVLAHRERFERRKT
Rv3154|M.tuberculosis_H37Rv ANANGNVEGLAALIFSRYLWAFELTSALLITAAVGAMVLAHRERFERRKT
MMAR_1474|M.marinum_M ANANGNVEGLAALIFSRYLWAFELTSALLITAAIGAMVLAHRERFERRKS
MUL_2468|M.ulcerans_Agy99 ANANGNVEGLAALIFSRYLWAFELTSALLITAAIGAMVLAHRERFERRKS
MAV_4042|M.avium_104 ANANGNVEGLAALIFSRYLWAFELTSALLITAAVGAMVLAHRERFERRKT
Mflv_4490|M.gilvum_PYR-GCK ANGGGNVEGLAALIFTKYLWAFELTGALLITAALGAMVLAHRERFQRRKT
Mvan_1876|M.vanbaalenii_PYR-1 ANEDGNVEGLAALIFTKYLWAFELTGALLITAALGAMVLAHRERFERRKT
MSMEG_2054|M.smegmatis_MC2_155 ANSGGNVEGLAALIFTRYLWAFELTSALLITAALGAMVLAHRERFERRKT
TH_1088|M.thermoresistible__bu ANSGGNVEGVAALVFTRYLWAFEITATLLITAALGAMVLAHRERFGRRKT
MAB_2143|M.abscessus_ATCC_1997 ANSRGNVEGLAERIFIDYLWAFELTGALLITATIGAMVLAHREKLGQTGG
** *****:* :* ******:*.:*****::*********:: :
Mb3178|M.bovis_AF2122/97 QRELSQERFRPGGHPTPLPNPGVYARHNAVDVAALLPDGSYSELSVPRML
Rv3154|M.tuberculosis_H37Rv QRELSQERFRPGGHPTPLPNPGVYARHNAVDVAALLPDGSYSELSVPRML
MMAR_1474|M.marinum_M QRELSQERFRPGGHPTPLPNPGVYARHNAVDVAALLPDGSYSELSVPGIL
MUL_2468|M.ulcerans_Agy99 QRELSQERFRPGGHPTPLPNPGVYARHNAVDVAALLPDGSYSELSVPGIL
MAV_4042|M.avium_104 QRELSQERFRAGGHPTPLPNPGVYARHNAVDVAALLPDGSYSKLSVSPIL
Mflv_4490|M.gilvum_PYR-GCK QRELAVERFAPGGHPTTLPNPGVYARHNAVDVPARLPDGSGSELSVSAIL
Mvan_1876|M.vanbaalenii_PYR-1 QRELAAERFAPGGHPTTLPNPGVYARHNAVDVPARLPDGSGSELSVSAIL
MSMEG_2054|M.smegmatis_MC2_155 QRELAIERFQTGGHPTPLPNPGVYARHNSVDVPARLPDGSESPLSVSTIL
TH_1088|M.thermoresistible__bu QRELAEERFREGGYPTTLPPPGVYATRNAVDVPARLPDGSDAEISVNPML
MAB_2143|M.abscessus_ATCC_1997 QRELAIRRFQHGDRATPLPNPGVYARHNAVDVPALLPDGSFSELSVSAVL
****: .** *. .*.** ***** :*:***.* ***** : :** :*
Mb3178|M.bovis_AF2122/97 RTRGADGLQTPSPGAVSGSLEGGAS
Rv3154|M.tuberculosis_H37Rv RTRGADGLQTPSPGAVSGSLEGGAS
MMAR_1474|M.marinum_M QTRGADGLQTPSPEAITG----GAS
MUL_2468|M.ulcerans_Agy99 QTRGADGLQTPSPEAITG----GAS
MAV_4042|M.avium_104 QSRGGDGKDVAAPEPSPSLSQGGAS
Mflv_4490|M.gilvum_PYR-GCK QVRPVPTDERS-PGGEE--------
Mvan_1876|M.vanbaalenii_PYR-1 QVRPVPSDSDSESAGERR-------
MSMEG_2054|M.smegmatis_MC2_155 PHRTVG----SATNGKR--------
TH_1088|M.thermoresistible__bu RPREEHE------------------
MAB_2143|M.abscessus_ATCC_1997 TPREMEAEVGRRES-----------
*