For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTEPTGDQTPEIIGVRRGMFGAKGSGDTSGYGRLVRPVALPGGSPRPYGGYFDEVVDRLAEAVGDDAFTE SIERVVIHRDELTLEVHRDRLVEVAQALRDDPALRFELCLGVSGVHYPDDTGRELHAAYPLMSITHNRRI RLEVAVPDDDPHIPSLFGVYPTVDWHERETYDFFGIIFDGHPSLTRIEMPDDWVGHPQRKDYPLGGIPVE YHGAEIPPPDQRRAYN
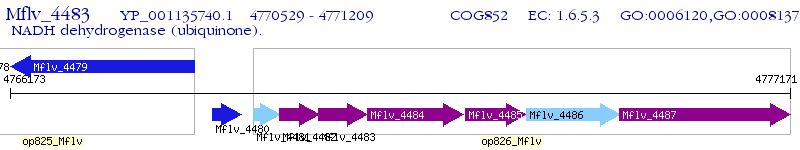
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4483 | - | - | 100% (226) | NADH dehydrogenase subunit C |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3171 | nuoC | 1e-103 | 76.82% (220) | NADH dehydrogenase subunit C |
| M. tuberculosis H37Rv | Rv3147 | nuoC | 1e-103 | 76.82% (220) | NADH dehydrogenase subunit C |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2136 | - | 3e-97 | 72.44% (225) | NADH dehydrogenase subunit C |
| M. marinum M | MMAR_1481 | nuoC | 1e-103 | 75.45% (224) | NADH dehydrogenase I (chain C) NuoC (NADH- ubiquinone |
| M. avium 104 | MAV_4035 | - | 1e-102 | 77.98% (218) | NADH dehydrogenase subunit C |
| M. smegmatis MC2 155 | MSMEG_2061 | - | 1e-108 | 80.27% (223) | NADH dehydrogenase subunit C |
| M. thermoresistible (build 8) | TH_0635 | nuoC | 1e-101 | 74.21% (221) | PROBABLE NADH DEHYDROGENASE I (CHAIN C) NUOC |
| M. ulcerans Agy99 | MUL_2461 | nuoC | 1e-104 | 75.89% (224) | NADH dehydrogenase subunit C |
| M. vanbaalenii PYR-1 | Mvan_1883 | - | 1e-115 | 84.82% (224) | NADH dehydrogenase subunit C |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2136|M.abscessus_ATCC_1997 MTGGEQKPN-------------EIIGVRRGLFGVRGSGDTSGYGGLVQPI
MSMEG_2061|M.smegmatis_MC2_155 MSTSNGSANGTNGVGLPRGDEPEIIAVRRGMFGNRDTGDTSGYGRLVRPV
TH_0635|M.thermoresistible__bu MSDHTPDHRE------PGRGDPEVIAVRRGMFGVGDSGDTSGYGGLVKPV
Mflv_4483|M.gilvum_PYR-GCK MTEPTGDQT------------PEIIGVRRGMFGAKGSGDTSGYGRLVRPV
Mvan_1883|M.vanbaalenii_PYR-1 MTGDD-EPD------------PEVIGVRRGMFGAKGSGDTSGYGRLIRSV
Mb3171|M.bovis_AF2122/97 MSPPNQDAQ----EGRPDSPTAEVVDVRRGMFGVSGTGDTSGYGRLVRQV
Rv3147|M.tuberculosis_H37Rv MSPPNQDAQ----EGRPDSPTAEVVDVRRGMFGVSGTGDTSGYGRLVRQV
MMAR_1481|M.marinum_M MSSPDQNPSDA--AGQTGSSNEEVVDVRRGMFGVKGTGDTSGYGRLVREI
MUL_2461|M.ulcerans_Agy99 MSSPDQNPSDA--AGQTGSSNEEVVDVRRGMFGVKGSGDTSGYGRLVREI
MAV_4035|M.avium_104 MSSPDQDPREA--IAQGD---DEVIDVRRGMFGAAGTGDTSGYGRLVRTV
*: . *:: ****:** .:******* *:: :
MAB_2136|M.abscessus_ATCC_1997 SLPVSSNRPYGGYFDQVVDRLESVLAEQENVTYADAVEGVVVYRDQLTIH
MSMEG_2061|M.smegmatis_MC2_155 ALPGSTPRPYGGYFDAVMDRLAEVLGEER---YAMSIERVVVYRDQLTIE
TH_0635|M.thermoresistible__bu AMPGAATRPYGGCFDAVVDALIAALGAQR---YAAAVERVVVYRDELTLD
Mflv_4483|M.gilvum_PYR-GCK ALPGGSPRPYGGYFDEVVDRLAEAVGDD---AFTESIERVVIHRDELTLE
Mvan_1883|M.vanbaalenii_PYR-1 ALPGSSPRPYGGYFDDVVDALESALGHD---VFAESVERVVVYRDELTLE
Mb3171|M.bovis_AF2122/97 VLPGSSPRPYGGYFDDIVDRLAEALRHER-VEFEDAVEKVVVYRDELTLH
Rv3147|M.tuberculosis_H37Rv VLPGSSPRPYGGYFDDIVDRLAEALRHER-VEFEDAVEKVVVYRDELTLH
MMAR_1481|M.marinum_M VLPGSSPRPYGFYFDEIADRLAEALNRDG-VEFEDAIEKVVVYRNELTLH
MUL_2461|M.ulcerans_Agy99 VLPGSSPRPYGFYFDEIADRLAEALNRDG-VEFEDAIEKVVVYRNELTLH
MAV_4035|M.avium_104 TLPGSSPRPYGSYFDDVVDTLTESLQSNG-IEFHQAIEKVVVYRDELTLH
:* .: **** ** : * * : : : ::* **::*::**:.
MAB_2136|M.abscessus_ATCC_1997 VKAEHLVQVAQSLRDDPQLRFELSLGVSGVHYPDDSARELHAVYPLMSIT
MSMEG_2061|M.smegmatis_MC2_155 VSRVQLPAVASVLRDDPDLRFELCLGVSGVHYPEDTGRELHAVYPLMSIT
TH_0635|M.thermoresistible__bu IRREQLRAVATALRDDPDLRFELCVGVSGVHYPEDTGRELHAVYGFLSIT
Mflv_4483|M.gilvum_PYR-GCK VHRDRLVEVAQALRDDPALRFELCLGVSGVHYPDDTGRELHAAYPLMSIT
Mvan_1883|M.vanbaalenii_PYR-1 VHRDRLVDVAQTLRDDPDLRFELCLGVSGVHYPDDADRELHAVYPLMSIT
Mb3171|M.bovis_AF2122/97 VRRDLLPRVAQRLRDEPELRFELCLGVSGVHYPHETGRELHAVYPLQSIT
Rv3147|M.tuberculosis_H37Rv VRRDLLPRVAQRLRDEPELRFELCLGVSGVHYPHETGRELHAVYPLQSIT
MMAR_1481|M.marinum_M VRREALLRVAQSLRDEPELRFELCLGVNGVHYPHETGRELHAVYPLQSIT
MUL_2461|M.ulcerans_Agy99 VRREALLRVAQSLRDEPELRFELCLGVNGVHYPHETGRELHAVYPLQSIT
MAV_4035|M.avium_104 VDRAALPHVAQHLRDDPRLRFEMCLGVSGVHYPHETGRELHAVYPLQSIT
: * ** ***:* ****:.:**.*****.:: *****.* : ***
MAB_2136|M.abscessus_ATCC_1997 WNRRIMLEVAVPESDPHIPSLNAVYPTTDWHERETYDFFGIIFDDHPALT
MSMEG_2061|M.smegmatis_MC2_155 HNRRIQLEVAAPDADPHIPSLYAVYPTTDWHERETYDFFGIIFDGHPSLT
TH_0635|M.thermoresistible__bu HNRRLRLEVAVPDADPHIPSLFGIYPTTDWHERETYDFFGIVFDGHPALT
Mflv_4483|M.gilvum_PYR-GCK HNRRIRLEVAVPDDDPHIPSLFGVYPTVDWHERETYDFFGIIFDGHPSLT
Mvan_1883|M.vanbaalenii_PYR-1 HNRRIRIEVAAPDSDPHIPSLFRVYPTTDWHERETYDFFGLIFDGHPALT
Mb3171|M.bovis_AF2122/97 HNRRLRLEVSAPDSDPHIPSLFAIYPTNDWHERETYDFFGIIFDGHPALT
Rv3147|M.tuberculosis_H37Rv HNRRLRLEVSAPDSDPHIPSLFAIYPTNDWHERETYDFFGIIFDGHPALT
MMAR_1481|M.marinum_M HNRRLRLEVSAPDSDPHIPSLYAVYPTNDWHERETYDFFGIIFDGHPSLT
MUL_2461|M.ulcerans_Agy99 HNRRLRLEVSAPDSDPHIPSLYAVYPTNDWHERETYDFFGIIFDGHPSLT
MAV_4035|M.avium_104 HNRRVRLEVAVPDGDPHIPSLYRIYPTTDWHERETYDFFGIIFDGHPSLT
***: :**:.*: ******* :*** ************::**.**:**
MAB_2136|M.abscessus_ATCC_1997 RIQMPDDWDGHPQRKDYPLGGVPVEYHGATIAPPDQRRSYN
MSMEG_2061|M.smegmatis_MC2_155 RIEMPDDWEGHPQRKDYPLGGIPVEYHGAQIPPPDQRRSYS
TH_0635|M.thermoresistible__bu RIAMPDDWEGHPQRKDYPLGGVPVQYHGATIPPPDERRAYN
Mflv_4483|M.gilvum_PYR-GCK RIEMPDDWVGHPQRKDYPLGGIPVEYHGAEIPPPDQRRAYN
Mvan_1883|M.vanbaalenii_PYR-1 RIEMPDDWVGHPQRKDYPLGGIPVEYHGAEIPPPDERRAYS
Mb3171|M.bovis_AF2122/97 RIEMPDDWQGHPQRKDYPLGGIPVEYKGAQIPPPDERRGYN
Rv3147|M.tuberculosis_H37Rv RIEMPDDWQGHPQRKDYPLGGIPVEYKGAQIPPPDERRGYN
MMAR_1481|M.marinum_M RIEMPDDWQGHPQRKDYPLGGIPVEYKGAQIPPPDERRGYN
MUL_2461|M.ulcerans_Agy99 RIEMPDDWQGHPQRKDYPLGGIPVEYKGAQIPPPDERRGYN
MAV_4035|M.avium_104 RIEMPDDWHGHPQRKDYPLGGIPVEYKGAQIPPPDERRAYN
** ***** ************:**:*:** *.***:**.*.