For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTGDMYFIPRAYPHHIENIGTDEWHFLIFFDQPFPADIGYRASASAYSREVLAAAFNTHIEDLPRFPFT PADPLIVSRINPVD
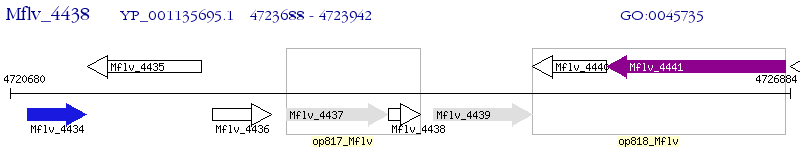
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4438 | - | - | 100% (84) | hypothetical protein Mflv_4438 |
| M. gilvum PYR-GCK | Mflv_4437 | - | 3e-06 | 40.58% (69) | cupin domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4766 | - | 6e-05 | 35.82% (67) | oxalate decarboxylase |
| M. marinum M | MMAR_2972 | - | 7e-37 | 78.57% (84) | putative sugar transport protein |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2254 | - | 2e-32 | 74.07% (81) | oxalate decarboxylase OxdC, putative |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1922 | - | 4e-45 | 94.05% (84) | cupin 2 domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4438|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1922|M.vanbaalenii_PYR-1 -MTTSVAGSSHATSLHDGEIVEESDLGSIRRVTADNLPILQGLSIKRVLL
MMAR_2972|M.marinum_M MTTTSDRLTTHAISLWDGEIVEESDLGSIRRVTADTFPILRGMSIKRVLI
MSMEG_2254|M.smegmatis_MC2_155 -MTTSLSHSIHATSLLDSELVEENDLGSIRRVTADNFPILRGLSIKRLVI
MAB_4766|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4438|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1922|M.vanbaalenii_PYR-1 NPGAMRTPHWHANANELTYCVSGTALVSMLDDYSRFSTFIVTAGQMFHAA
MMAR_2972|M.marinum_M NPGAMRTPHWHANANELTYCVSGTSLVSVLDSGSQFSSFTVSAGEMFHID
MSMEG_2254|M.smegmatis_MC2_155 NPGAMRTPHWHANANELTYCVSGSALVSVLDTASRFSTFTVSAGEMFHVD
MAB_4766|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4438|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1922|M.vanbaalenii_PYR-1 SGSLHHIENIGEDVAEFIIAFRHERPEDFGLGATLGAFSDAVLGNSYDLP
MMAR_2972|M.marinum_M SGSLHHIENIGEDVAEFIIAFRHERPEDFGLGAAFGAMTDAVLGNTYDLD
MSMEG_2254|M.smegmatis_MC2_155 SGSLHHIENIGTEPAEFIITFRNERPEDFGLGAAFGAMTDAVLGNTYDLD
MAB_4766|M.abscessus_ATCC_1997 -------------------MKSINRRTVLGGVGVAGVAAAAGAGTDWAFS
Mflv_4438|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1922|M.vanbaalenii_PYR-1 ASDFARIHRDTTDHKLAARTGPPDVPAAAYFNDPHKFDIEAQAPGLNYVS
MMAR_2972|M.marinum_M AAAFGTIRRRASDRALAARSGPAVVPGTAWYNDPHKFAVEAQQPPISVAV
MSMEG_2254|M.smegmatis_MC2_155 ASDFAALRRDTTDRALAARRGDPVIPQAAHFGDPHKFGVEAMTPPVTSAV
MAB_4766|M.abscessus_ATCC_1997 PPRSHDNPEDVTVTDDAARFGDPRLPAELNTAQPHLFHLGALAP-QTFDG
Mflv_4438|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1922|M.vanbaalenii_PYR-1 GSARFARDQFWPVLK--DMSMYSLRVSEDGMREPHWHPVTAEMGYVQHGD
MMAR_2972|M.marinum_M GSARLARVQYWPALK--DLSMYSLRIKEDGMREPHWHPVTAEMGYVQQGS
MSMEG_2254|M.smegmatis_MC2_155 GSARTARVQFWPALK--DLSMYSLRVREDGMREPHWHPVTAEMGYVQSGS
MAB_4766|M.abscessus_ATCC_1997 GDLRQAHEGNFPILTGQQASVVMVTLQPGGIREPHWHPSAWEINVITSGV
Mflv_4438|M.gilvum_PYR-GCK ----------------MTTGDMYFIPRAYPHHIENIGTDEWHFLIFFDQP
Mvan_1922|M.vanbaalenii_PYR-1 ARMTVMNPDGTLDTWTMSAGDMYFIPRAYPHHIENIGTDEWHFLIFFDQP
MMAR_2972|M.marinum_M ARMTVMDPDGTLDTWHLNAGDVYFIPRAYPHHIEVIDCDEWHFTIFFDQP
MSMEG_2254|M.smegmatis_MC2_155 ARMTVMDPDGTLDTWELQRGDVYFIPRAYPHHIEVVHAPDLHFLIFFDQP
MAB_4766|M.abscessus_ATCC_1997 ATWTLLDPMGHSETFDAHVGDVVFAPQGSLHYFENKGPDDLKLLIVFNAS
**: * *:. *::* : :: *.*: .
Mflv_4438|M.gilvum_PYR-GCK FP---ADIGYRASASAYSREVLAAAFNTHIEDLPRFPFTPADPLIVSRIN
Mvan_1922|M.vanbaalenii_PYR-1 FP---ADIGYRASASAYSREVLAAAFNTHIDDLPRFPFTPADPLIVARTN
MMAR_2972|M.marinum_M TP---GDIGYRASVSAYSREVLAATFNTHIDDLPDFPFTRTDPLVVNRIN
MSMEG_2254|M.smegmatis_MC2_155 TP---ADVGYRTSVSAYSREVLAATFDTHIDDLPDFPLTTADPLIVGRRN
MAB_4766|M.abscessus_ATCC_1997 TAEGKDDIGIGASISKLPPDVLSAVFGVPTETFAKFKKINESVTILRRPQ
. *:* :* * . :**:*.*.. : :. * . :: * :
Mflv_4438|M.gilvum_PYR-GCK PVD------
Mvan_1922|M.vanbaalenii_PYR-1 PVD------
MMAR_2972|M.marinum_M PVDRHLVGE
MSMEG_2254|M.smegmatis_MC2_155 PLDR-----
MAB_4766|M.abscessus_ATCC_1997 T--------
.