For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTDVVIGERDVEVTLRPTARLLTCPCGKRQPSVYDRRRRRWRHLDLGVKRLWLVNEIRRLNCPDCGVITE AVPWARPGSRFTRDFEDMVLWLAQRSDRTTVSTLMRCAWESVTAIINRGVAELLDKRRLQTLYRIGVDEI CYRHPHRYLTIIGDHDTGTVVNIRPGRSEQSLTDFYTAQPDSMLKQIQAVSMDVSKPYIGATRSKVPEAI ICFDGFHIMRWINRALDAVFSEAAAGPDKVHMSSAQWRTTRWALRTGENRLTDTKRELVNQIAKQNRRVG RAWALKEQARDLYRYDHQPGVARALLKAWITAAKRCRIPAFVALGKRLAVYFDGILAAIELGISNALAES INAKIRLINARGYGHHTAETLTSMIYLCLGGLQVKLPTRT
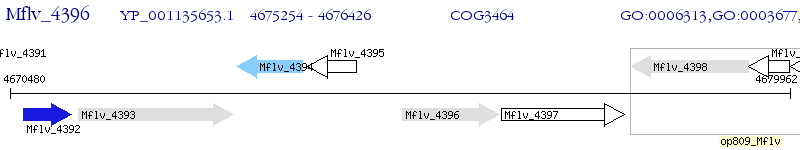
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4396 | - | - | 100% (390) | transposase, IS204/IS1001/IS1096/IS1165 family protein |
| M. gilvum PYR-GCK | Mflv_2896 | - | 0.0 | 100.00% (390) | transposase, IS204/IS1001/IS1096/IS1165 family protein |
| M. gilvum PYR-GCK | Mflv_1834 | - | 0.0 | 100.00% (390) | transposase, IS204/IS1001/IS1096/IS1165 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3827 | - | 8e-27 | 44.59% (157) | transposase |
| M. tuberculosis H37Rv | Rv3798 | - | 7e-56 | 35.32% (402) | transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1440 | - | 4e-09 | 24.72% (267) | ISL3 family transposase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6697 | - | 5e-17 | 24.58% (419) | IS1096, tnpA protein |
| M. thermoresistible (build 8) | TH_3774 | - | 7e-16 | 24.43% (348) | PUTATIVE transposase IstA protein, putative |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1440|M.marinum_M --------------------------------------------------
MSMEG_6697|M.smegmatis_MC2_155 --------------------------------------------------
Mb3827|M.bovis_AF2122/97 --------------------------------------------------
Rv3798|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4396|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu MLTWEDDVEVHALAKRGWTISAIARHTGFDRKTVRKYLAGDGTPGVRARP
MMAR_1440|M.marinum_M --------------------------------------------------
MSMEG_6697|M.smegmatis_MC2_155 --------------------------------------------------
Mb3827|M.bovis_AF2122/97 --------------------------------------------------
Rv3798|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4396|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu GPDPFDPFVDYVTARLTEDPHLWARTLFDELEDLGFGLSYQSLTRNIRTR
MMAR_1440|M.marinum_M --------------------------------------------------
MSMEG_6697|M.smegmatis_MC2_155 --------------------------------------------------
Mb3827|M.bovis_AF2122/97 --------------------------------------------------
Rv3798|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4396|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu NLRPVCEACRTATGRPNAVIPHPPGEETQWDWLELPDPPASWGWGKTAHL
MMAR_1440|M.marinum_M --------------------------------------------------
MSMEG_6697|M.smegmatis_MC2_155 --------------------------------------------------
Mb3827|M.bovis_AF2122/97 --------------------------------------------------
Rv3798|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4396|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu LVGSLSHSGKWRGNLAPSEDQPHLVAGLDRVTRGLGGLTRVWRFDRMATV
MMAR_1440|M.marinum_M --------------------------------------------------
MSMEG_6697|M.smegmatis_MC2_155 --------------------------------------------------
Mb3827|M.bovis_AF2122/97 --------------------------------------------------
Rv3798|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4396|M.gilvum_PYR-GCK --------------------------------------------------
TH_3774|M.thermoresistible__bu CDPGSGRVTASFAGVAKHYGVSVAICPPRRGNRKGVVEKVNHTAAQRWWR
MMAR_1440|M.marinum_M MPDAT----FACPDLTTFCRLD-ELGLEATGQHLATDRAVLACRITDDGS
MSMEG_6697|M.smegmatis_MC2_155 MPDATGRAGFACADLTTFCRLD-ELGLEVTGQRLDPDRAVLACRVADEDR
Mb3827|M.bovis_AF2122/97 --MRNVRLFRALLGVDKRTVIE-DIEFEEDDAGDGARVIARVRPRSAVLR
Rv3798|M.tuberculosis_H37Rv --MRNVRLFRALLGVDKRTVIE-DIEFEEDDAGDGARVIARVRPRSAVLR
Mflv_4396|M.gilvum_PYR-GCK ----------------------------MTDVVIGERDVEVTLRPTARLL
TH_3774|M.thermoresistible__bu TLADDLTVEAAQASLDRFARVRGDTRLRATADGRSSVAVVAXIYVRTAEQ
MMAR_1440|M.marinum_M WCRRCG------------EQGTVRDGVT--RQLAHEPFGWRPTTLLVTIR
MSMEG_6697|M.smegmatis_MC2_155 WCRRCG------------EEGVVRDSVT--RTLAHEPFGWRPTALLVTIR
Mb3827|M.bovis_AF2122/97 RCGRCG------------RKASWYDRGAGLRQWRSLDWGTVEVFLEAEAP
Rv3798|M.tuberculosis_H37Rv RCGRCG------------RKASWYDRGAGLRQWRSLDWGTVEVFLEAEAP
Mflv_4396|M.gilvum_PYR-GCK TCPCGK------------RQPSVYDRRR--RRWRHLDLGVKRLWLVNEIR
TH_3774|M.thermoresistible__bu WVGVCPGCMVRSGVCPGCMVRSARSKGWVSTRPRDIKIGPDIPRIVWRKR
. * :
MMAR_1440|M.marinum_M RYRCQ--GCAHVWRQDSGKAAEPRAKLSRRGLRWALEAIVCRHLSVARIA
MSMEG_6697|M.smegmatis_MC2_155 RYRCA--GCAHVWRQDASAAAEPRARLSRRALRWALEALVCQHLSVARVA
Mb3827|M.bovis_AF2122/97 RVNCP--THGPTVVAVPWARHHAGHTYAFDDT----VAWLAVACSKTAVC
Rv3798|M.tuberculosis_H37Rv RVNCP--THGPTVVAVPWARHHAGHTYAFDDT----VAWLAVACSKTAVC
Mflv_4396|M.gilvum_PYR-GCK RLNCP--DCGVITEAVPWARPGSRFTRDFEDM----VLWLAQRSDRTTVS
TH_3774|M.thermoresistible__bu KWVCANMCCDRKSFTESVPSIPPRARVTVRAKREMASAVLDDDRSVKAVA
: * . . : . :.
MMAR_1440|M.marinum_M EALAVSWNTANNAVLAEGRRVLIADPARFDSVAVIG--VDEHVWRHTRRG
MSMEG_6697|M.smegmatis_MC2_155 EALAVSWNTANNAVLAEGQRVLIADPARFDGVAVIG--VDEHVWRHTRRG
Mb3827|M.bovis_AF2122/97 ELMRIAWRTVG-AIVARVWADTEKRIDRFANLRRIG--IDEISYKRHHR-
Rv3798|M.tuberculosis_H37Rv ELMRIAWRTVG-AIVARVWADTEKRIDRFANLRRIG--IDEISYKRHHR-
Mflv_4396|M.gilvum_PYR-GCK TLMRCAWESVT-AIINRGVAELLD-KRRLQTLYRIG--VDEICYRHPHR-
TH_3774|M.thermoresistible__bu AAYGCTWNTCHDAVIATADPVLAGEPEAVSVLGIDETRRGKAKWETCSQT
:*.: *:: . : .: :. :
MMAR_1440|M.marinum_M DKYVIVIIDLTPVRDKTGPARLLDMVEGRSKKAFQDWLAGRPKQWRDGVE
MSMEG_6697|M.smegmatis_MC2_155 DKYVTVIIDLTPVRDGTGPARLLDMVEGRSKKAFADWLAQRPQEWRDRVD
Mb3827|M.bovis_AF2122/97 --YLTVVVDHD-------SGRLVWAAPSHPGLVL----------------
Rv3798|M.tuberculosis_H37Rv --YLTVVVDHD-------SGRLVWAAPGHDKATLGLFFDALGAERAAQIT
Mflv_4396|M.gilvum_PYR-GCK --YLTIIGDHD-------TGTVVNIRPGRSEQSLTDFYTAQPDSMLKQIQ
TH_3774|M.thermoresistible__bu GTRSWVDRFDTGLVDITGTGGLLVQVNGRAAKPVTDWLDQRDPGWKATIE
: .. :: .: .
MMAR_1440|M.marinum_M VVAMDGFSGFKTATTEELPGAATVMAPFHVVRLAGNALDECRRRVQMDTC
MSMEG_6697|M.smegmatis_MC2_155 VVAMDGFSGFKTAATEELPDAATVMDPFHVVRLAGNALDECRRRVQLATC
Mb3827|M.bovis_AF2122/97 ----------------RCPGR-----------------------------
Rv3798|M.tuberculosis_H37Rv HVSADAADWIADVVTERCPDAIQCADPFHVVAWATEALDVERRRAWNDAR
Mflv_4396|M.gilvum_PYR-GCK AVSMDVSKPYIGATRSKVPEAIICFDGFHIMRWINRALDAVFSEAAAGPD
TH_3774|M.thermoresistible__bu FVAIDMSVTYANAARQALPHAKLIVDRFHLVKRANEMVDEVRRRTTQAHR
*
MMAR_1440|M.marinum_M GHRGRRTDPLYA----------------------CRRTLQTGADLLTDKQ
MSMEG_6697|M.smegmatis_MC2_155 GHRGRSTDPLYR----------------------SRRTLHTGADLLTDRQ
Mb3827|M.bovis_AF2122/97 --------------------------------------------------
Rv3798|M.tuberculosis_H37Rv --AIARTEPKWGRGRPGKNAAPRPGRERARRLKGARYALWKNPEDLTERQ
Mflv_4396|M.gilvum_PYR-GCK --KVHMSSAQWR---------------------TTRWALRTGENRLTDTK
TH_3774|M.thermoresistible__bu LRRGRKSDPEWIN----------------------RRRLLRAAERLTDEQ
MMAR_1440|M.marinum_M KTRLTALLS---CAVG------------FVLRLNSADPGAGFAMLLAACV
MSMEG_6697|M.smegmatis_MC2_155 KARLAALFA---ANAHAEIEATWAMYQRTVAAYREPDRTKGRTMMAALIT
Mb3827|M.bovis_AF2122/97 --------------------------------------------------
Rv3798|M.tuberculosis_H37Rv SAKLAWIAK---TDPRLYRAYLLKESLRHVFSVK-GEE--GKQALDRWIS
Mflv_4396|M.gilvum_PYR-GCK RELVNQIAK---QNRRVGRAWALKEQARDLYRYD-HQPGVARALLKAWIT
TH_3774|M.thermoresistible__bu RHKLFVKLTSADPNGDIAAAWIAKELLRDVLACT-ARGGLTYEIRDALYR
MMAR_1440|M.marinum_M AISR-------------------------------------FACDGCHSV
MSMEG_6697|M.smegmatis_MC2_155 TLSTGVPTSLTELITLGRTLKKRAADVLAYFDRPGTSNGPTEAINGRLEH
Mb3827|M.bovis_AF2122/97 --------------------------------------------------
Rv3798|M.tuberculosis_H37Rv WAQR---CRIPVFVELAARIKRHRVAIDAALDHG-LSQGLIESTNTKIRL
Mflv_4396|M.gilvum_PYR-GCK AAKR---CRIPAFVALGKRLAVYFDGILAAIELG-ISNALAESINAKIRL
TH_3774|M.thermoresistible__bu FYTFCAACSVPEISKLARTISAWQEPMILAIQTG-LSNARSEGYNRIIKH
MMAR_1440|M.marinum_M AFGSAWTDDLLPGWW-------------------------
MSMEG_6697|M.smegmatis_MC2_155 LRGSALGFRNLTNYIARSLLETGGFRTQLRQPRR------
Mb3827|M.bovis_AF2122/97 ----------------------------------------
Rv3798|M.tuberculosis_H37Rv LTRIAFGFRSPQALIALAMLTLAGHRPTLPGRHNHPQISQ
Mflv_4396|M.gilvum_PYR-GCK INARGYGHHTAETLTSMIYLCLGGLQVKLPTRT-------
TH_3774|M.thermoresistible__bu VGRIAFGFRNPENQRRRVRWACTRQSRRTPPSTKQVRPC-