For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDEVLARAAIFQDVDPGAVAALCSQLQKVSFPRGHRVFNEGDLGDTLYIITSGRAKIGCTSPDGRESLLT LMGPSDMFGELAIFDPGPRTSSVTALTALHAVMMDRDALRFWIAERPEIAEQLLRVLARRLRRTNDALSD LIFTDVPGRVAKQLLDLARRFGVQQEGSLRVDHGLTQEEIAQLAGSSRETINKALADFVQRGWIEQRGRT TIIHDPARLARRAQQ
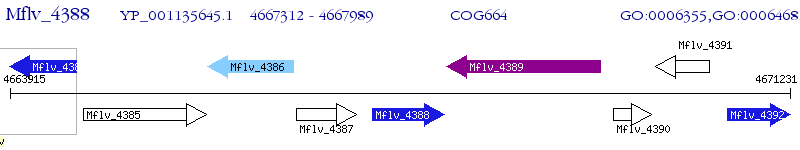
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4388 | - | - | 100% (225) | CRP/FNR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1371 | - | 1e-89 | 72.77% (224) | CRP/FNR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1915 | - | 1e-08 | 27.56% (127) | cyclic nucleotide-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3700 | - | 2e-90 | 72.77% (224) | CRP/FNR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3676 | - | 2e-90 | 72.77% (224) | CRP/FNR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02302 | - | 1e-91 | 73.66% (224) | putative Crp/Fnr-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0416c | - | 1e-90 | 73.21% (224) | Crp/Fnr family transriptional regulator |
| M. marinum M | MMAR_5164 | - | 2e-91 | 73.66% (224) | regulatory protein |
| M. avium 104 | MAV_0453 | - | 1e-90 | 73.21% (224) | transcriptional regulator, Crp/Fnr family protein |
| M. smegmatis MC2 155 | MSMEG_6189 | - | 6e-91 | 74.11% (224) | transcriptional regulator, Crp/Fnr family protein |
| M. thermoresistible (build 8) | TH_2129 | - | 1e-91 | 74.55% (224) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_4250 | - | 5e-91 | 73.21% (224) | regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5435 | - | 2e-90 | 73.21% (224) | CRP/FNR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_02302|M.leprae_Br4923 MDEILARAGIFQGVEPSAVAALTKQLQPVDFPRGHTVFTEGEPGDRLYII
MAV_0453|M.avium_104 MDEILARAGIFQGVEPSAVTALTKQLQPVDFPRGHTVFAEGEPGDRLYII
MMAR_5164|M.marinum_M MDEILARAGIFQGVEPGAVAALTKQLQPVDFPRGHTVFAEGEPGDRLYII
MUL_4250|M.ulcerans_Agy99 MDEILARAGIFQGVEPGAVAALTKQLQPVDFPRGHTVFAEGEPGDRLYII
MAB_0416c|M.abscessus_ATCC_199 MDEILARAGIFQGVEPSAVSALTKQLQPVDFPRGHTVFAEGEPGDRLYII
Mvan_5435|M.vanbaalenii_PYR-1 MDEILARAGIFQGVEPSAVSALTKQLQPVDFPRGHTVFAEGEPGDRLYII
MSMEG_6189|M.smegmatis_MC2_155 MDEILARAGIFQGVEPTAVAALTKQLQPVDFPRGHTVFAEGEPGDRLYII
Mb3700|M.bovis_AF2122/97 MDEILARAGIFQGVEPSAIAALTKQLQPVDFPRGHTVFAEGEPGDRLYII
Rv3676|M.tuberculosis_H37Rv MDEILARAGIFQGVEPSAIAALTKQLQPVDFPRGHTVFAEGEPGDRLYII
TH_2129|M.thermoresistible__bu VDEILARAGIFQGVEPSAVAALTKQLQPVDFPRGHTVFAEGEPGDRLYII
Mflv_4388|M.gilvum_PYR-GCK MDEVLARAAIFQDVDPGAVAALCSQLQKVSFPRGHRVFNEGDLGDTLYII
:**:****.***.*:* *::** .*** *.***** ** **: ** ****
MLBr_02302|M.leprae_Br4923 VSGKVKIGRRSPDGRENLLTIMGPSDMFGELSIFDPGPRTSSATTITEVR
MAV_0453|M.avium_104 VSGKVKIGRRSPDGRENLLTIMGPSDMFGELSIFDPGPRTSSATTITEVR
MMAR_5164|M.marinum_M VAGKVKIGRRSPDGRENLLTIMGPSDMFGELSIFDPGPRTSSATTITEVR
MUL_4250|M.ulcerans_Agy99 VAGKVKIGRRSPDGRENLLTIMGPSDMFGELSIFDPGPRTSSATTITEVR
MAB_0416c|M.abscessus_ATCC_199 ITGKVKIGRRSPDGRENLLTIMGPSDMFGELSIFDPGPRTSSATTITEVR
Mvan_5435|M.vanbaalenii_PYR-1 ISGKVKIGRRSPDGRENLLTIMGPSDMFGELSIFDPGPRTSSATTITEVR
MSMEG_6189|M.smegmatis_MC2_155 ISGKVKIGRRSPDGRENLLTIMGPSDMFGELSIFDPGPRTSSATTITEVR
Mb3700|M.bovis_AF2122/97 ISGKVKIGRRAPDGRENLLTIMGPSDMFGELSIFDPGPRTSSATTITEVR
Rv3676|M.tuberculosis_H37Rv ISGKVKIGRRAPDGRENLLTIMGPSDMFGELSIFDPGPRTSSATTITEVR
TH_2129|M.thermoresistible__bu ISGKVKIGRRSPDGRENLLTIMGPSDMFGELSIFDPGPRTSSATTITEVR
Mflv_4388|M.gilvum_PYR-GCK TSGRAKIGCTSPDGRESLLTLMGPSDMFGELAIFDPGPRTSSVTALTALH
:*:.*** :*****.***:**********:**********.*::* ::
MLBr_02302|M.leprae_Br4923 AVSMDRDALRAWIADRPEIAEQLLRVLARRLRRTNNNLADLIFTDVPGRV
MAV_0453|M.avium_104 AVSMDRDALRAWIADRPEIAEQLLRVLARRLRRTNNNLADLIFTDVPGRV
MMAR_5164|M.marinum_M AVSMDRDALRAWIADRPEIAEQLLRVLARRLRRTNNNLADLIFTDVPGRV
MUL_4250|M.ulcerans_Agy99 AVSMDRDALRAWIADRPEIAEQLLRVLARRLRRTSNNLADLIFTDVPGRV
MAB_0416c|M.abscessus_ATCC_199 AVSMDRDALRAWIADRPEIAEQLLRVLARRLRRTNNNLADLIFTDVPGRV
Mvan_5435|M.vanbaalenii_PYR-1 AVSMDREALRAWIADRPEIAEQLLRVLARRLRRTNNNLADLIFTDVPGRV
MSMEG_6189|M.smegmatis_MC2_155 AVSMDRDALRAWIADRPEIAEQLLRVLARRLRRTNNNLADLIFTDVPGRV
Mb3700|M.bovis_AF2122/97 AVSMDRDALRSWIADRPEISEQLLRVLARRLRRTNNNLADLIFTDVPGRV
Rv3676|M.tuberculosis_H37Rv AVSMDRDALRSWIADRPEISEQLLRVLARRLRRTNNNLADLIFTDVPGRV
TH_2129|M.thermoresistible__bu AVSMDRDALRGWIKDRPEIAEQLLRVLARRLRRTNNALADLIFTDVPGRV
Mflv_4388|M.gilvum_PYR-GCK AVMMDRDALRFWIAERPEIAEQLLRVLARRLRRTNDALSDLIFTDVPGRV
** ***:*** ** :****:**************.: *:***********
MLBr_02302|M.leprae_Br4923 AKQLLQLAQRFGTQEGGAMRVTHDLTQEEIAQLVGASRETVNKALADFAH
MAV_0453|M.avium_104 AKQLLQLAQRFGTQEGGAMRVTHDLTQEEIAQLVGASRETVNKALADFAH
MMAR_5164|M.marinum_M AKQLLQLAQRFGTQEGGAMRVTHDLTQEEIAQLVGASRETVNKALADFAH
MUL_4250|M.ulcerans_Agy99 AKQLLQLAQRFGTQEGGAMRVTHDLTQEEIAQLVGASRETVNKALADFAH
MAB_0416c|M.abscessus_ATCC_199 AKQLLQLAQRFGTQEGGALRVTHDLTQEEIAQLVGASRETVNKALADFAH
Mvan_5435|M.vanbaalenii_PYR-1 AKQLLQLAQRFGTQEGGALRVTHDLTQEEIAQLVGASRETVNKALADFAH
MSMEG_6189|M.smegmatis_MC2_155 AKQLLQLAQRFGTQEGGALRVTHDLTQEEIAQLVGASRETVNKALADFAH
Mb3700|M.bovis_AF2122/97 AKQLLQLAQRFGTQEGGALRVTHDLTQEEIAQLVGASRETVNKALADFAH
Rv3676|M.tuberculosis_H37Rv AKQLLQLAQRFGTQEGGALRVTHDLTQEEIAQLVGASRETVNKALADFAH
TH_2129|M.thermoresistible__bu AKQLLQLAQRFGTQEGGMLRVTHDLTQEEIAQLVGASRETVNKALADFAQ
Mflv_4388|M.gilvum_PYR-GCK AKQLLDLARRFGVQQEGSLRVDHGLTQEEIAQLAGSSRETINKALADFVQ
*****:**:***.*: * :** *.*********.*:****:*******.:
MLBr_02302|M.leprae_Br4923 RGWIRLDGKSVLISDSERLARRAR-
MAV_0453|M.avium_104 RGWIRLEGKSVLISDSERLARRAR-
MMAR_5164|M.marinum_M RGWIRLEGKSVLISDSERLARRAR-
MUL_4250|M.ulcerans_Agy99 RGWIRLEGKSVLISDSERLARRAR-
MAB_0416c|M.abscessus_ATCC_199 RGWIRLEGKSVLISDSERLARRAR-
Mvan_5435|M.vanbaalenii_PYR-1 RGWIRLEGKSVLISDSERLARRAR-
MSMEG_6189|M.smegmatis_MC2_155 RGWIRLEGKSVLISDSERLARRAR-
Mb3700|M.bovis_AF2122/97 RGWIRLEGKSVLISDSERLARRAR-
Rv3676|M.tuberculosis_H37Rv RGWIRLEGKSVLISDSERLARRAR-
TH_2129|M.thermoresistible__bu RGWIRLEGKSVLISDSERLARRAR-
Mflv_4388|M.gilvum_PYR-GCK RGWIEQRGRTTIIHDPARLARRAQQ
****. *::.:* *. ******: