For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SPEAIPDPESMLAEQAHDALDPDLRRSWQELADEVREHQFRYYIRDAPIITDAEFDQLLRRLQALEDEYP ELRTPDSPTQLVGGAGFATDFTAAEHLERMLSLDNVFDLEELAAWAARIRTEIGADAQYLCELKIDGVAL ALVYRDGRLERAATRGDGRTGEDVTLNARTIEDIPERLSGTNEFPLPTVVEVRGEVFFRVADFEDLNAGL VAEGKPPFANPRNSAAGSLRQKNPAVTARRRLRMICHGLGHVESSGGSPFPTLHDAYRALKAWGLPVSDH TAQVTGLDAVTERIAYWGEHRHDVEHEIDGVVVKVDAVALQRRLGATSRAPRWAVAYKYPPEEAQTRLLD IRVNVGRTGRVTPFAYMEPVKVAGSTVGLATLHNASEVKRKGVLIGDTVVIRKAGDVIPEVLGPVVDLRD GSEREFVFPTHCPECGSELAPAKEGDADIRCPNTRSCPAQLRERVFHVAGRGAFDIEGLGYEAATALLQA GVIADEGDLFGLTADDLLRTELFTTKAGELSANGKRLLANLGKAKAQPLWRVLVALSIRHVGPTAARALA TEFGSLEAIETASEEQLAGTEGVGPTIAAAVIDWFTVDWHRAIVDKWREAGVRMADERDASIERTLEGLS IVVTGSLAGFSRDEAKEAIIARGGKAAGSVSKKTAYVVAGDAPGSKYDKAIELGVPVLDEDGFRRLLAEG PERDAEDGEPG*
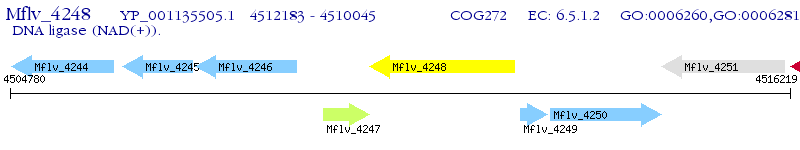
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4248 | ligA | - | 100% (712) | NAD-dependent DNA ligase LigA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3039c | ligA | 0.0 | 80.12% (684) | NAD-dependent DNA ligase LigA |
| M. tuberculosis H37Rv | Rv3014c | ligA | 0.0 | 80.12% (684) | NAD-dependent DNA ligase LigA |
| M. leprae Br4923 | MLBr_01705 | ligA | 0.0 | 76.90% (684) | NAD-dependent DNA ligase LigA |
| M. abscessus ATCC 19977 | MAB_3345c | ligA | 0.0 | 79.77% (682) | NAD-dependent DNA ligase LigA |
| M. marinum M | MMAR_1694 | ligA | 0.0 | 79.74% (691) | NAD-dependent DNA ligase LigA |
| M. avium 104 | MAV_3868 | ligA | 0.0 | 80.58% (690) | NAD-dependent DNA ligase LigA |
| M. smegmatis MC2 155 | MSMEG_2362 | ligA | 0.0 | 86.32% (702) | NAD-dependent DNA ligase LigA |
| M. thermoresistible (build 8) | TH_0630 | ligA | 0.0 | 88.02% (384) | PROBABLE DNA LIGASE |
| M. ulcerans Agy99 | MUL_1933 | ligA | 0.0 | 80.00% (685) | NAD-dependent DNA ligase LigA |
| M. vanbaalenii PYR-1 | Mvan_2112 | ligA | 0.0 | 89.90% (703) | NAD-dependent DNA ligase LigA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4248|M.gilvum_PYR-GCK MSPEAIPDPESMLAEQAHDALDPDLRRSWQELADEVREHQFRYYIRDAPI
Mvan_2112|M.vanbaalenii_PYR-1 MSSKATQDPEAVLAEQSDDATEAGLRRQWQELADEVRDHQFRYYVRDAPI
MSMEG_2362|M.smegmatis_MC2_155 MSEKATGEVEAELPEHP----DADERRRWQELADEVREHQFRYYVKDAPI
MAV_3868|M.avium_104 MSSAEADSVP------------PEVRRQWQELAETVREHQFRYYIKDAPI
TH_0630|M.thermoresistible__bu --------------------------------------------------
MAB_3345c|M.abscessus_ATCC_199 --------------------MDSDIQRQWGELAEEVRGHQFRYYVKDAPV
Mb3039c|M.bovis_AF2122/97 MSSPDADQTA------------PEVLRQWQALAEEVREHQFRYYVRDAPI
Rv3014c|M.tuberculosis_H37Rv MSSPDADQTA------------PEVLRQWQALAEEVREHQFRYYVRDAPI
MMAR_1694|M.marinum_M --------MT------------PEVLRQWQELAEQVREHQFRYYVRDAPV
MUL_1933|M.ulcerans_Agy99 ------------------------MLRQWQELAEQVREHQFRYYVRDAPV
MLBr_01705|M.leprae_Br4923 MSSPEADPIG------------PEVLQQWRKLTEEVREHQFRYYVRDAPI
Mflv_4248|M.gilvum_PYR-GCK ITDAEFDQLLRRLQALEDEYPELRTPDSPTQLVGGAGFATDFTAAEHLER
Mvan_2112|M.vanbaalenii_PYR-1 ITDAEFDAMLRRLEALEEAHPELRTPDSPTQLVGGAGFATDFTAAEHLER
MSMEG_2362|M.smegmatis_MC2_155 ISDAEFDKLLRELQALEDAHPELRTPDSPTQLVGGAGFATEFAPAEHLER
MAV_3868|M.avium_104 ISDAEFDALFNELLALEERHPELRVADSPTQLVGGAGFATDFAEAAHLER
TH_0630|M.thermoresistible__bu --------------------------------------------------
MAB_3345c|M.abscessus_ATCC_199 ISDGQFDELLRRLTALEEQYPELRTPDSPTQLVGGAGFVTEFRSVDHLER
Mb3039c|M.bovis_AF2122/97 ISDAEFDELLRRLEALEEQHPELRTPDSPTQLVGGAGFATDFEPVDHLER
Rv3014c|M.tuberculosis_H37Rv ISDAEFDELLRRLEALEEQHPELRTPDSPTQLVGGAGFATDFEPVDHLER
MMAR_1694|M.marinum_M ITDAEFDELLRRLEALEEQYPELRTPDSPTQLVGGAGFATEFEPVEHLER
MUL_1933|M.ulcerans_Agy99 ITDAEFDELLRRLEALEEQYPELRTPDSPTQLVGGAGFATEFEPVEHLER
MLBr_01705|M.leprae_Br4923 ISDAEFDALLDRLTVLEEQHPELCTPDSPTQLVGGAGFMTDFVATEHLER
Mflv_4248|M.gilvum_PYR-GCK MLSLDNVFDLEELAAWAARIRTEIGADAQYLCELKIDGVALALVYRDGRL
Mvan_2112|M.vanbaalenii_PYR-1 MLSLDNVFTPDELAAWAARIRTEIGADAQYLCELKIDGVALSLVYRDGRL
MSMEG_2362|M.smegmatis_MC2_155 MLSLDNVFDSDELTAWAARISSETGDAAHFLCELKIDGVALSLVYRDGRL
MAV_3868|M.avium_104 MLSLDDVFDTDELIAWSNRVENEIGKDPHYLCELKIDGVALSLVYRDGRL
TH_0630|M.thermoresistible__bu --------------------------------------------------
MAB_3345c|M.abscessus_ATCC_199 MLSLDNAFSSDELTAWDARVRGDIGQEPEYLCELKIDGVALSLVYENGVL
Mb3039c|M.bovis_AF2122/97 MLSLDNAFTADELAAWAGRIHAEVGDAAHYLCELKIDGVALSLVYREGRL
Rv3014c|M.tuberculosis_H37Rv MLSLDNAFTADELAAWAGRIHAEVGDAAHYLCELKIDGVALSLVYREGRL
MMAR_1694|M.marinum_M MLSLDNAFNTEELTAWAGRIHADVGDSAAYLCELKIDGVALSLVYEGGRL
MUL_1933|M.ulcerans_Agy99 MLSLDNAFNTEELTAWAGRIHADVGDSAAYLCELKIDGVALSLVYEGGRL
MLBr_01705|M.leprae_Br4923 MFSLDNAFSGDELNAWVTRIRAEIGNDAHYLCELKIDGVALSLVYRDGRL
Mflv_4248|M.gilvum_PYR-GCK ERAATRGDGRTGEDVTLNARTIEDIPERLSGTNEFPLPTVVEVRGEVFFR
Mvan_2112|M.vanbaalenii_PYR-1 ERGATRGDGRTGEDVTLNARTIEDVPEKLTGTEEFPLPSVLEVRGEVFFR
MSMEG_2362|M.smegmatis_MC2_155 ERGATRGDGRTGEDVTLNARTIDDIPERLTPSDEFPVPAVLEVRGEVFFR
MAV_3868|M.avium_104 ERAATRGDGRVGEDVTLNARTIDDVPERLSPSDDFPVPALLEVRGEVFFL
TH_0630|M.thermoresistible__bu --------------------------------------------------
MAB_3345c|M.abscessus_ATCC_199 VRGATRGDGRSGEDVTLNARTIEDVPERLAKSEKYPIPALLEVRGEVFFR
Mb3039c|M.bovis_AF2122/97 TRASTRGDGRTGEDVTLNARTIADVPERLTPGDDYPVPEVLEVRGEVFFR
Rv3014c|M.tuberculosis_H37Rv TRASTRGDGRTGEDVTLNARTIADVPERLTPGDDYPVPEVLEVRGEVFFR
MMAR_1694|M.marinum_M TRASTRGDGRTGEDVTLNARTIEDVPERLSHSEDHRMPEVLEVRGEVFFR
MUL_1933|M.ulcerans_Agy99 TRASTRGDGRTGEDVTLNARTIEDVPERLSHSEDHRMPEVLEVRGEVFFR
MLBr_01705|M.leprae_Br4923 TRATTRGDGRTGEDVTLNARTIEDVPERLTVSDIYPLPEVLEVRGEVFFR
Mflv_4248|M.gilvum_PYR-GCK VADFEDLNAGLVAEGKPPFANPRNSAAGSLRQKNPAVTARRRLRMICHGL
Mvan_2112|M.vanbaalenii_PYR-1 IADFEDLNAGLVAEGKPPFANPRNSAAGSLRQKNPAVTARRRLRMICHGL
MSMEG_2362|M.smegmatis_MC2_155 VADFEELNAGLVAEGKPPFANPRNSAAGSLRQKNPAVTARRKLRMICHGI
MAV_3868|M.avium_104 LADFEALNASLVEDGKAPFANPRNSAAGSLRQKNPAVTARRKLRMICHGI
TH_0630|M.thermoresistible__bu --------------------------------------------------
MAB_3345c|M.abscessus_ATCC_199 LEDFEALNASLVEESKPPFANPRNSAAGSLRQKNPAITARRRLRMICHGL
Mb3039c|M.bovis_AF2122/97 LDDFQALNASLVEEGKAPFANPRNSAAGSLRQKDPAVTARRRLRMICHGL
Rv3014c|M.tuberculosis_H37Rv LDDFQALNASLVEEGKAPFANPRNSAAGSLRQKDPAVTARRRLRMICHGL
MMAR_1694|M.marinum_M VADFQALNASLVEEGKAPFANPRNSAAGSLRQKDPAVTARRRLRMICHGL
MUL_1933|M.ulcerans_Agy99 VADFQALNASLVEEGKAPFANPRNSAAGSLRQKDPAVTARRRLRMICHGL
MLBr_01705|M.leprae_Br4923 VVDFQALNTSLVEEGKNPFANPRNSAAGSLRQKDPAVTAHRKLRMICHGL
Mflv_4248|M.gilvum_PYR-GCK GHVESSGGSPFPTLHDAYRALKAWGLPVSDHTAQVTGLDAVTERIAYWGE
Mvan_2112|M.vanbaalenii_PYR-1 GHIEDAAGFPFRTLHDAYRALQAWGLPVSPHTAQVTGLDAVTERIAYWGE
MSMEG_2362|M.smegmatis_MC2_155 GYTEG---FTPASLHDAYRALGAWGLPVSEHTTKVSTVAEVAERIAYWGE
MAV_3868|M.avium_104 GRTEG---FSPKTQHEAYTALSVWGLPVAEQTARVRGLAAVQERIGYWGE
TH_0630|M.thermoresistible__bu --------------------------------------------------
MAB_3345c|M.abscessus_ATCC_199 GRAEG---FSPESLHDAYLALGEWGLPVSTHTTKVRGIAKVQERVNYWAE
Mb3039c|M.bovis_AF2122/97 GHVEG---FRPATLHQAYLALRAWGLPVSEHTTLATDLAGVRERIDYWGE
Rv3014c|M.tuberculosis_H37Rv GHVEG---FRPATLHQAYLALRAWGLPVSEHTTLATDLAGVRERIDYWGE
MMAR_1694|M.marinum_M GHTEG---FRPATLHQAYLALQAWGLPVSQHTTLVADLAGVQERIDYWGE
MUL_1933|M.ulcerans_Agy99 GHTEG---FRPATLHQAYLALQAWGLPVSQHTTLVADLAEVQARIDYWGE
MLBr_01705|M.leprae_Br4923 GHIEG---FRPATQHQAYLALRDWGLPVSEHTTLVNDIAGVQGCIAYWAE
Mflv_4248|M.gilvum_PYR-GCK HRHDVEHEIDGVVVKVDAVALQRRLGATSRAPRWAVAYKYPPEEAQTRLL
Mvan_2112|M.vanbaalenii_PYR-1 HRHDVEHEIDGVVVKVDEVALQRRLGATSRAPRWAVAYKYPPEEAQTKLL
MSMEG_2362|M.smegmatis_MC2_155 HRHDVEHEIDGVVVKVDEVALQRRLGATSRAPRWAVAYKYPPEEATTKLL
MAV_3868|M.avium_104 HRYELQHEIDGVVVKVDDVALQRRLGATSRAPRWAVAYKYPPQEAQTKLL
TH_0630|M.thermoresistible__bu ------------------VALQRRLGATSRAPRWAVAYKYPPEEAQTRLL
MAB_3345c|M.abscessus_ATCC_199 HRHDVEHEIDGVVVKVDTVALQRRLGSTSRAPRWAIAYKYPPEEATTELL
Mb3039c|M.bovis_AF2122/97 HRHEVDHEIDGVVVKVDEVALQRRLGSTSRAPRWAIAYKYPPEEAQTKLL
Rv3014c|M.tuberculosis_H37Rv HRHEVDHEIDGVVVKVDEVALQRRLGSTSRAPRWAIAYKYPPEEAQTKLL
MMAR_1694|M.marinum_M HRHEVDHEIDGVVVKVDDVALQRRLGSTSRAPRWAIAYKYPPEEAQTKLL
MUL_1933|M.ulcerans_Agy99 HRHEVDHEIDGVVVKVDDVALQRRLGSTSRAPRWAIAYKYPPEEAQTKLL
MLBr_01705|M.leprae_Br4923 HRHEVDHEIDGIVVKIDEVTLQRRLGSTSRAPRWAIAYKYLPEEAQTKLL
*:******:********:**** *:** *.**
Mflv_4248|M.gilvum_PYR-GCK DIRVNVGRTGRVTPFAYMEPVKVAGSTVGLATLHNASEVKRKGVLIGDTV
Mvan_2112|M.vanbaalenii_PYR-1 DIRVNVGRTGRVTPFAYMEPVKIAGSTVGLATLHNGSEVKRKGVLIGDTV
MSMEG_2362|M.smegmatis_MC2_155 DIRVNVGRTGRVTPFAYMEPVKVAGSTVGLATLHNASEVKRKGVLIGDTV
MAV_3868|M.avium_104 DIRVNVGRTGRVTPFAFMTPVKVAGSTVGLATLHNAAEVKRKGVLIGDTV
TH_0630|M.thermoresistible__bu DIRVNVGRTGRVTPFAYMEPVKVAGSTVSLATLHNAAEVKRKGVLIGDTV
MAB_3345c|M.abscessus_ATCC_199 DIRVSVGRTGRVTPFAYMTPVKVAGSTVSLATLHNASEVKRKGVLIGDTV
Mb3039c|M.bovis_AF2122/97 DIRVNVGRTGRITPFAFMTPVKVAGSTVGQATLHNASEIKRKGVLIGDTV
Rv3014c|M.tuberculosis_H37Rv DIRVNVGRTGRITPFAFMTPVKVAGSTVGQATLHNASEIKRKGVLIGDTV
MMAR_1694|M.marinum_M DIRVNVGRTGRVTPFAFMTPVKVAGSTVAQATLHNASEVKRKGVLIGDTV
MUL_1933|M.ulcerans_Agy99 DIRVNVGRTGRVTPFAFMTPVKVAGSTVAQATLHNASEVKRKGVLIGDTV
MLBr_01705|M.leprae_Br4923 DIRVNVGRTGRVTPFAFMTPVKVAGSTVGQATLHNPSEVKRKGVLIGDTV
****.******:****:* ***:*****. ***** :*:***********
Mflv_4248|M.gilvum_PYR-GCK VIRKAGDVIPEVLGPVVDLRDGSEREFVFPTHCPECGSELAPAKEGDADI
Mvan_2112|M.vanbaalenii_PYR-1 VIRKAGDVIPEVLGPVVDLRDGSEREFVMPTHCPECGTELAPAKEGDADI
MSMEG_2362|M.smegmatis_MC2_155 VIRKAGDVIPEVLGPVVDLRDGTEREFEFPTHCPECGTELAPAKEGDADI
MAV_3868|M.avium_104 MIRKAGDVIPEVLGPVVDLRDGTEREFVMPTTCPECGTPLAPAKEGDADI
TH_0630|M.thermoresistible__bu MIRKAGDVIPEVLGPVVELRDGTEREFVMPTECPECGTPLAPEKEGDADI
MAB_3345c|M.abscessus_ATCC_199 VIRKAGDVIPEVLGPVADLRNGNEREFVMPTACPECGTTLAHEKEGDADI
Mb3039c|M.bovis_AF2122/97 VIRKAGDVIPEVLGPVVELRDGSEREFIMPTTCPECGSPLAPEKEGDADI
Rv3014c|M.tuberculosis_H37Rv VIRKAGDVIPEVLGPVVELRDGSEREFIMPTTCPECGSPLAPEKEGDADI
MMAR_1694|M.marinum_M VIRKAGDVIPEVLGPVVDLRDGSEREFVMPTTCPECGTPLAPEKEGDADI
MUL_1933|M.ulcerans_Agy99 VIRKAGDVIPEVLGPVVDLRDGSEREFVMPTTCPECGTPLAPEKEGDADI
MLBr_01705|M.leprae_Br4923 VIRKAGDVIPEVLGPVVDLRDGSEREFVMPTTCPECGTTLAPEKEGDADI
:***************.:**:*.**** :** *****: ** *******
Mflv_4248|M.gilvum_PYR-GCK RCPNTRSCPAQLRERVFHVAGRGAFDIEGLGYEAATALLQAGVIADEGDL
Mvan_2112|M.vanbaalenii_PYR-1 RCPNSRTCPAQLRERVFHVAGRGAFDIEGLGYEAAIALLQARVITDEGDL
MSMEG_2362|M.smegmatis_MC2_155 RCPNSRSCPAQLRERLFHLAGRGAFDIEGLGYEAATALLAAQVIPDEGDL
MAV_3868|M.avium_104 RCPNARSCPAQLRERVFHVAGRGALDIEGLGYEAATALLKAGVIADEGDL
TH_0630|M.thermoresistible__bu RCPNSRSCPAQLRERVFHVASRGALDIEGLGYEAATALLEAGVIADEGDL
MAB_3345c|M.abscessus_ATCC_199 RCPNSRSCPAQLRERVFHVAGRGAFDIEALGYEAAIALLAAGVIEDEGDL
Mb3039c|M.bovis_AF2122/97 RCPNARGCPGQLRERVFHVASRNGLDIEVLGYEAGVALLQAKVIADEGEL
Rv3014c|M.tuberculosis_H37Rv RCPNARGCPGQLRERVFHVASRNGLDIEVLGYEAGVALLQAKVIADEGEL
MMAR_1694|M.marinum_M RCPNTRSCPGQLRERVFHVSSRNALDIEMLGYEAGAALLSARVIGDEGDL
MUL_1933|M.ulcerans_Agy99 RCPNTRSCPGQLRERVFHVSSRNALDIEMLGYEAGAALLSARVIGDEGDL
MLBr_01705|M.leprae_Br4923 RCPNARSCPGQLRERVFHVASRSALDIQGLGYEAGVALLAAQVITSEGDL
****:* **.*****:**::.*..:**: *****. *** * ** .**:*
Mflv_4248|M.gilvum_PYR-GCK FGLTADDLLRTELFTTKAGELSANGKRLLANLGKAKAQPLWRVLVALSIR
Mvan_2112|M.vanbaalenii_PYR-1 FTLTADDLLRTDLFTTKAGELSANGKRLLTNLGKAKAQPLWRVLVALSIR
MSMEG_2362|M.smegmatis_MC2_155 FTLTADDLLRTELFTTKKGELSANGKRLLANLTKAKEQPLWRVLVALSIR
MAV_3868|M.avium_104 FALTEDDLLRTELFRTKAGTLSANGTRLLQNLQKAKKVALWRVLVALSIR
TH_0630|M.thermoresistible__bu FDLTEDDLLRTEFFTTKNGALSANGKRLLANIAKARNQPLWRVLVALSIR
MAB_3345c|M.abscessus_ATCC_199 FGLTADDLLRTDLFKTKSGALSANGARLLDNLDKAKQQPLWRVLVALSIR
Mb3039c|M.bovis_AF2122/97 FALTERDLLRTDLFRTKAGELSANGKRLLVNLDKAKAAPLWRVLVALSIR
Rv3014c|M.tuberculosis_H37Rv FALTERDLLRTDLFRTKAGELSANGKRLLVNLDKAKAAPLWRVLVALSIR
MMAR_1694|M.marinum_M FGLTEEELLRTDLFRTKAGDLSANGRRLLANLDKAKAAPLWRVLVALSIR
MUL_1933|M.ulcerans_Agy99 FGLTEEELLRTDLFRTKAGDLSANGRRLLANLDKAKAAPLWRVLVALSIR
MLBr_01705|M.leprae_Br4923 FTLTEKALLRTELFRNKAGELSANGKRLLVNVDKAKTAPLWRVLVALSIR
* ** ****::* .* * ***** *** *: **: .***********
Mflv_4248|M.gilvum_PYR-GCK HVGPTAARALATEFGSLEAIETASEEQLAGTEGVGPTIAAAVIDWFTVDW
Mvan_2112|M.vanbaalenii_PYR-1 HVGPTAARALATEFGSLDAIVAASEEELAAVEGVGPTIAAAVTDWFTVDW
MSMEG_2362|M.smegmatis_MC2_155 HVGPTAARALATEFGSLEAIEAASEEELAAVEGVGPTIAAAVKDWFTVDW
MAV_3868|M.avium_104 HVGPTAARALATEFGDLDSIMSASTERLAAVEGVGPTIAAALTEWFTVDW
TH_0630|M.thermoresistible__bu HVGPTAARALATEFGSLDAIIAASEQELAAVDGVGPTIAAAVKEWFTVDW
MAB_3345c|M.abscessus_ATCC_199 HVGPTAARALATEFGDLEAIEGASVEQLAAVEGVGATIAAAVVDWFSVDW
Mb3039c|M.bovis_AF2122/97 HVGPTAARALATEFGSLDAIAAASTDQLAAVEGVGPTIAAAVTEWFAVDW
Rv3014c|M.tuberculosis_H37Rv HVGPTAARALATEFGSLDAIAAASTDQLAAVEGVGPTIAAAVTEWFAVDW
MMAR_1694|M.marinum_M HVGPTAARALATEFGSIDAIVSATTEQLAAVEGVGPTIASAVSEWFTVDW
MUL_1933|M.ulcerans_Agy99 HVGPTAARALATEFGSIDAIVAATTEQLAAVEGVGPTIASAVSEWFTVDW
MLBr_01705|M.leprae_Br4923 HVGPTAARALATEFGSVDAILAASPEQLAAVEGVGTTIAAAVTEWFTVDW
***************.:::* *: :.**..:***.***:*: :**:***
Mflv_4248|M.gilvum_PYR-GCK HRAIVDKWREAGVRMADERDASIERTLEGLSIVVTGSLAGFSRDEAKEAI
Mvan_2112|M.vanbaalenii_PYR-1 HRTIVDKWRAAGVRMADERDASIERTLDGLSIVVTGSLTGYSRDEAKEAI
MSMEG_2362|M.smegmatis_MC2_155 HRAIVDKWRAAGVRMADERDASIERTLEGLSIVVTGSLAGFSRDQAKEAI
MAV_3868|M.avium_104 HRAIVDKWRGAGVRMADERDASVPRTLEGLTVVVTGSLAGFSRDDAKEAI
TH_0630|M.thermoresistible__bu HRTIVDKWRAAGVRMADERDTSIERTLEGVTVVVTGSLTGFSRDEAKEAI
MAB_3345c|M.abscessus_ATCC_199 HRAIVDKWRAAGVRTADERDDSIPRNLEGLSIVVTGSLPGFSRDEAKEAI
Mb3039c|M.bovis_AF2122/97 HREIVDKWRAAGVRMVDERDESVPRTLAGLTIVVTGSLTGFSRDDAKEAI
Rv3014c|M.tuberculosis_H37Rv HREIVDKWRAAGVRMVDERDESVPRTLAGLTIVVTGSLTGFSRDDAKEAI
MMAR_1694|M.marinum_M HREIVEKWRAAGVRMADERDDSVPRTLAGVTVVVTGSLPGFSRDEAKEAI
MUL_1933|M.ulcerans_Agy99 HREIVERWRAAGVRMADERDDSVPRTLAGVTVVVTGSLPGFSRDEAKEAI
MLBr_01705|M.leprae_Br4923 HRVIVNKWRAAGVRMVDERDTSVLPTCEGLTIVVTGSLAGFSRDDAKEAI
** **::** **** .**** *: . *:::******.*:***:*****
Mflv_4248|M.gilvum_PYR-GCK IARGGKAAGSVSKKTAYVVAGDAPGSKYDKAIELGVPVLDEDGFRRLLAE
Mvan_2112|M.vanbaalenii_PYR-1 IARGGKAAGSVSKKTAYVVAGDSPGSKYDKAIELGVPVLDEDGFRRLLEN
MSMEG_2362|M.smegmatis_MC2_155 IARGGKAAGSVSKKTAYVVAGDAPGSKYDKAVELGVPVLDEDGFRRLLEQ
MAV_3868|M.avium_104 LARGGKAAGSVSKKTDYVVAGDSPGSKYDKAVELGVPILDEDGFRKLLAE
TH_0630|M.thermoresistible__bu IARGGKASGSVSKKTDFVVAGDSPGSKYDKAVELGVPILDEDGFRRLLAE
MAB_3345c|M.abscessus_ATCC_199 IARGGKSASSVSKKTAFVVVGDSPGSKYDKAVELGVTILDEDGFRALLAD
Mb3039c|M.bovis_AF2122/97 VARGGKAAGSVSKKTNYVVAGDSPGSKYDKAVELGVPILDEDGFRRLLAD
Rv3014c|M.tuberculosis_H37Rv VARGGKAAGSVSKKTNYVVAGDSPGSKYDKAVELGVPILDEDGFRRLLAD
MMAR_1694|M.marinum_M VTRGGKAAGSVSKKTSYVVAGDAPGSKYDKAVELGVPILDEDGFRKLLEQ
MUL_1933|M.ulcerans_Agy99 VTRGGKAAGSVSKKTSYVVAGDAPGSKYDKAVELGVPILDEDGFRKLLEQ
MLBr_01705|M.leprae_Br4923 VARGGKVAGSVSKKIAYVVVGDLPGYKYDKAVELGVPILNEDGFRLLLEQ
::**** :.***** :**.** ** *****:****.:*:***** ** :
Mflv_4248|M.gilvum_PYR-GCK GPERDAEDGEPG
Mvan_2112|M.vanbaalenii_PYR-1 GP--DTPDS---
MSMEG_2362|M.smegmatis_MC2_155 GPPVEPAE----
MAV_3868|M.avium_104 GPPETPAD----
TH_0630|M.thermoresistible__bu GPAGV-------
MAB_3345c|M.abscessus_ATCC_199 GPPA--------
Mb3039c|M.bovis_AF2122/97 GPASRT------
Rv3014c|M.tuberculosis_H37Rv GPASRT------
MMAR_1694|M.marinum_M GPPAEVGEPT--
MUL_1933|M.ulcerans_Agy99 GPPAEAGEPT--
MLBr_01705|M.leprae_Br4923 GPPVQQVVD---
**