For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGRPVHTFEVVRTEQLTPHVIRVVLGGNGFDTFTPGDHTDSYVKLVFVADDVDVSSLEQPLTLDSFNALP AQHRPTVRTYTVRKADPERREITIDFVVHGDHGVAGPWAASATPGRRLFVMGPSGAYSPDPAADWHLFAG DESAVPAISAALESLPANAIGKVFLEVAGPEDEIELQAPAGVEVSWIYRGSRADLAPEESAGDHAPLIDA VKETPWLPGQVQVFIHGEAQAVMHNLRSYIRKERGVDATWASSISGYWRRGRTEETFREWKRELAAAEAK *
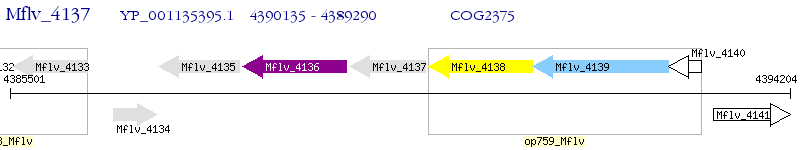
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4137 | - | - | 100% (281) | siderophore-interacting protein |
| M. gilvum PYR-GCK | Mflv_3144 | - | 3e-13 | 31.35% (185) | siderophore-interacting protein |
| M. gilvum PYR-GCK | Mflv_2227 | - | 3e-11 | 29.33% (208) | FAD-binding 9, siderophore-interacting domain-containing |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2919c | viuB | 1e-121 | 72.14% (280) | mycobactin utilization protein ViuB |
| M. tuberculosis H37Rv | Rv2895c | viuB | 1e-121 | 72.14% (280) | mycobactin utilization protein ViuB |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3205c | - | 7e-98 | 62.82% (277) | hypothetical protein MAB_3205c |
| M. marinum M | MMAR_1815 | viuB | 1e-122 | 72.14% (280) | mycobactin utilization protein ViuB |
| M. avium 104 | MAV_3748 | - | 1e-122 | 72.50% (280) | siderophore utilization protein |
| M. smegmatis MC2 155 | MSMEG_2511 | - | 1e-130 | 76.31% (287) | siderophore utilization protein |
| M. thermoresistible (build 8) | TH_0844 | - | 1e-116 | 71.22% (271) | POSSIBLE MYCOBACTIN UTILIZATION PROTEIN VIUB |
| M. ulcerans Agy99 | MUL_2063 | viuB | 1e-122 | 71.79% (280) | mycobactin utilization protein ViuB |
| M. vanbaalenii PYR-1 | Mvan_2205 | - | 1e-149 | 89.32% (281) | siderophore-interacting protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4137|M.gilvum_PYR-GCK MAGRPVHTFEVVRTEQLTPHVIRVVLGGN------GFDTFTPGDHTDSYV
Mvan_2205|M.vanbaalenii_PYR-1 MAGRPVHTFEVVRTEQLAPHIVRVVLGGN------GFDTFTPIEYTDSYV
MSMEG_2511|M.smegmatis_MC2_155 MAGRPIHTFAVVRREELTPHMVRLVLGGNNAGSGTGFGTFKPSEFSDSYV
Mb2919c|M.bovis_AF2122/97 MAGRPLHAFEVVATRHLAPHMVRVVLGGS------GFDTFVPSDFTDSYI
Rv2895c|M.tuberculosis_H37Rv MAGRPLHAFEVVATRHLAPHMVRVVLGGS------GFDTFVPSDFTDSYI
MAV_3748|M.avium_104 MAGRPLQSFEVVRTEQLTPHMVRVVLRGK------DFDAFVPSKFTDSYV
MMAR_1815|M.marinum_M MAGRPVHTFEVVGTQQLAPHMVRVRVGGS------GFDTFVPSEFTDSYV
MUL_2063|M.ulcerans_Agy99 MAGRPVHTFEVVGTQQLAPHMVRVRVGGS------GFDTFVPSEFTDSYV
TH_0844|M.thermoresistible__bu ----------VVRTEEVSPHIVRVVLGGR------GFDTFTPNEFTDAYV
MAB_3205c|M.abscessus_ATCC_199 -MTRPLHELQVVRTEQVSPHITRVVLGGN------GFEGFEVSSFTDSYA
** ..::**: *: : * .* * ..:*:*
Mflv_4137|M.gilvum_PYR-GCK KLVFVADDVDVSSLEQPLTLDSFNALPAQHRPTVRTYTVRKADPERREIT
Mvan_2205|M.vanbaalenii_PYR-1 KLVFVGDDVDVSSLEQPLTLDSFNALPPQHRPVVRTYTVRRADPERREIT
MSMEG_2511|M.smegmatis_MC2_155 KLVIVPEGVDVSSLPQPLTLDSFQGLPEAQRPTVRTYTVRNVDPERGEIT
Mb2919c|M.bovis_AF2122/97 KLVFVDDDVDVGRLPRPLTLDSFADLPTAKRPPVRTMTVRHVDAAAREIA
Rv2895c|M.tuberculosis_H37Rv KLVFVDDDVDVGRLPRPLTLDSFADLPTAKRPPVRTMTVRHVDAAAREIA
MAV_3748|M.avium_104 KLAFVADDVDVAGLPQPLTLDSFAELPAEKKPSVRTLTVRHVDAASRQIA
MMAR_1815|M.marinum_M KLVFVAHDVDVAALPRPLTLDCFSSLPPEKQPAVRTLTVRRVDAAAREIV
MUL_2063|M.ulcerans_Agy99 KLVFVAHDVDVAALPRPLTLDCFSSLPPEKQPAVRTLTVRRVDAAAREIV
TH_0844|M.thermoresistible__bu KIIFVRDNIDVAALPQPLTQDSFKSLPPEQQPPIRTYTVRRVDRDRREIW
MAB_3205c|M.abscessus_ATCC_199 KLIFLPQSHEG---PGPRDIDEARDM----QATVRTYTIRSVDTDRREVA
*: :: .. : * * : :. :** *:* .* ::
Mflv_4137|M.gilvum_PYR-GCK IDFVVHG-DHGVAGPWAASATPGRRLFVMGPSGAYSPDPAADWHLFAGDE
Mvan_2205|M.vanbaalenii_PYR-1 VDFVVHG-EHGVAGPWAASATPGQRLFVMGPSGAYAPDPAADWYLFAGDE
MSMEG_2511|M.smegmatis_MC2_155 IDFVVHG-ELGVAGPWAASAAPGQPAYLMGPSGAFSPDPAADWYLLAGDE
Mb2919c|M.bovis_AF2122/97 VDIVLHG-EHGVAGPWAAGAQRGQPIYLMGPGGAYAPDPAADWHLLAGDE
Rv2895c|M.tuberculosis_H37Rv VDIVLHG-EHGVAGPWAAGAQRGQPIYLMGPGGAYAPDPAADWHLLAGDE
MAV_3748|M.avium_104 LDVVVHG-EHGIAGQWAATAQPGQPIYLMGPGGAYTPDPAADWHLLAGDE
MMAR_1815|M.marinum_M LDVVAHG-EHGVAGQWAAAAQPGEPVYLMGPSGAYKPDPEADWHLLAGDE
MUL_2063|M.ulcerans_Agy99 LDVVAHG-EHGVAGQWAAAAQPGEPVYLMGPNGAYKPDPEADWHLLAGDE
TH_0844|M.thermoresistible__bu IDIVIHGEEHGVAGPWVQAAQPGDPVYLMGPSGAYAPRPDADWHLFAGDE
MAB_3205c|M.abscessus_ATCC_199 IDFVVHG-DTGVAGPWASRAKPGDKLYITDSHGAYAPDPAADWHLLAGDE
:*.* ** : *:** *. * * :: .. **: * * ***:*:****
Mflv_4137|M.gilvum_PYR-GCK SAVPAISAALESLPANAIGKVFLEVAGPEDEIELQAPAGVEVSWIYRGSR
Mvan_2205|M.vanbaalenii_PYR-1 SALPAISAALESLPDNAIGKVFIEVAGSEDEIELKAPAGVDVSWIHRGGR
MSMEG_2511|M.smegmatis_MC2_155 AAIPAISTALEALPDNAIGKAFIEVAGPDDELELKAPAGVEIKWIYRGGR
Mb2919c|M.bovis_AF2122/97 SAIPAIAAALEALPPDAIGRAFIEVAGPDDEIGLTAPDAVEVNWVYRGGR
Rv2895c|M.tuberculosis_H37Rv SAIPAIAAALEALPPDAIGRAFIEVAGPDDEIGLTAPDAVEVNWVYRGGR
MAV_3748|M.avium_104 SALPAIAAALEALPPSAVGKAFIEVAGHEDEIPLTAPDGVEVHWVYRGGR
MMAR_1815|M.marinum_M SALPAIAAALEALPADAVGKAFIEVAGPDDEIELRAPESVQINWVYRGGR
MUL_2063|M.ulcerans_Agy99 SALPAIAAALEALPADAVGKAFIEVAGPDDEIELRAPESVQINWVYRGGR
TH_0844|M.thermoresistible__bu SALPAISASLEALPPDAVGYAFIEISGPEDEIPLAAPEGVEVRWIYRGGR
MAB_3205c|M.abscessus_ATCC_199 SAIPAISVALQALPEKARAKVFVEVAGPQDEIDFDTRGDVDVVWVHRGAG
:*:***:.:*::** .* . .*:*::* :**: : : *:: *::**.
Mflv_4137|M.gilvum_PYR-GCK ADLAPEESAGDHAPLIDAVKETPWLPGQVQVFIHGEAQAVMHNLRSYIRK
Mvan_2205|M.vanbaalenii_PYR-1 ADLAPEDSAGDHAPLIAAVKEAPWLPGQVQVFIHGEAQAVMHNLRPYIRK
MSMEG_2511|M.smegmatis_MC2_155 ADLVPEDRAGDHAPLIDAVKETPWLPGQVQVFIHGEAQAVMHNLRPYIRK
Mb2919c|M.bovis_AF2122/97 ADLVPEDRAGDHAPLIEAVTTTAWLPGQVHVFIHGEAQAVMHNLRPYVRN
Rv2895c|M.tuberculosis_H37Rv ADLVPEDRAGDHAPLIEAVTTTAWLPGQVHVFIHGEAQAVMHNLRPYVRN
MAV_3748|M.avium_104 ADLVPEDRAGDHAPLIEAVTSAPWLPGQVHVFIHGEAQAVMHNLRPYVRK
MMAR_1815|M.marinum_M ADLVSADRAGDHAPLIEAVKTALWLPGQVHVFIHGEAQAVMHNLRPYIRK
MUL_2063|M.ulcerans_Agy99 ADLVSADRAGDHAPLIEAVKTALWLPGQVHVFIHGEAQAVMHNLRPYIRK
TH_0844|M.thermoresistible__bu ADLVPDDQAGDNAPLIAAVKESPWLPGQVHVFIHGEAQAVMHNLRPYIRK
MAB_3205c|M.abscessus_ATCC_199 ADEVGEEHAGDNAPLVAAVRSAEWPPGQVQVFIHGEAQTVMHNLRPYIRK
** . : ***:***: ** : * ****:********:******.*:*:
Mflv_4137|M.gilvum_PYR-GCK ERGVDATWASSISGYWRRGRTEETFREWKRELAAAEAK-------
Mvan_2205|M.vanbaalenii_PYR-1 ERGVDARWASSISGYWRRGRTEETFRQWKRELAEAEAK-------
MSMEG_2511|M.smegmatis_MC2_155 ERGVEAKWAASISGYWRRGRTEETFRQWKAELAKAEAEATA----
Mb2919c|M.bovis_AF2122/97 ERGVDAKWASSISGYWRRGRTEEMFRKWKKELAEAEAGTH-----
Rv2895c|M.tuberculosis_H37Rv ERGVDAKWASSISGYWRRGRTEEMFRKWKKELAEAEAGTH-----
MAV_3748|M.avium_104 ERGVDAKWAASISGYWRRGRTEETFRQWKKELAQAESAQA-----
MMAR_1815|M.marinum_M ERGVDAKWASSISGYWRRGRTEETFRQWKRELAESEAGS------
MUL_2063|M.ulcerans_Agy99 ERGVDAKWASSISGYWRRGRTEETFRQWKRELAESEAGS------
TH_0844|M.thermoresistible__bu ERGVPAEWAS-ISGYWRRGRTEETFRQWKRELAEAEAAAATEVVG
MAB_3205c|M.abscessus_ATCC_199 ERGVAPKWAASISGYWRRGRTEETFRVWKQELAAAEATVKE----
**** . **: ************ ** ** *** :*: