For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ANYTAADVKRLRELTGAGMMDSKNALVEAEGDFDKAVELLRIKGAKDVGKRAERATAEGLVAAKDGALIE LNSETDFVAKNAEFQAVAEQIVAAAAAAKATDVDALKAAKLGDTTVEQTIADLSAKIGEKLELRRATYFD GQVETYLHKRAADLPPAVGVLVEYTGDDKSAAHAVALQIAALKAKYLTREDVPEDIVANERRIAEETARA EGKPEQALTKIVEGRVTGFYKDVVLLDQPSVSDNKKSVKALLDEAGVTVTRFARFEVGQA*
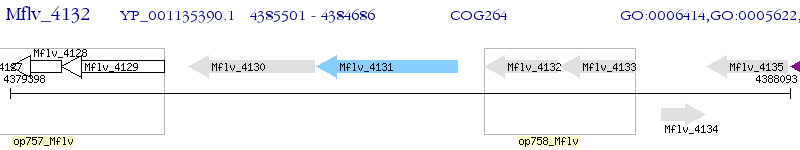
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4132 | tsf | - | 100% (271) | elongation factor Ts |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2913c | tsf | 1e-122 | 82.66% (271) | elongation factor Ts |
| M. tuberculosis H37Rv | Rv2889c | tsf | 1e-123 | 83.03% (271) | elongation factor Ts |
| M. leprae Br4923 | MLBr_01597 | tsf | 1e-114 | 76.45% (276) | elongation factor Ts |
| M. abscessus ATCC 19977 | MAB_3195c | tsf | 1e-121 | 83.39% (277) | elongation factor Ts |
| M. marinum M | MMAR_1820 | tsf | 1e-120 | 81.55% (271) | elongation factor Tsf |
| M. avium 104 | MAV_3743 | tsf | 1e-125 | 83.27% (275) | elongation factor Ts |
| M. smegmatis MC2 155 | MSMEG_2520 | tsf | 1e-133 | 90.55% (275) | elongation factor Ts |
| M. thermoresistible (build 8) | TH_0149 | tsf | 1e-128 | 85.45% (275) | PROBABLE ELONGATION FACTOR TSF (EF-TS) |
| M. ulcerans Agy99 | MUL_2068 | tsf | 1e-120 | 81.55% (271) | elongation factor Ts |
| M. vanbaalenii PYR-1 | Mvan_2210 | tsf | 1e-138 | 94.10% (271) | elongation factor Ts |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1820|M.marinum_M MANFTAADVKRLRELTGAGMLACKNALAESDGDFDKAVEALRIKGAKDVG
MUL_2068|M.ulcerans_Agy99 MANFTAADVKRLRELTGAGMLACKNALAESDGDFDKAVEALRIKGAKDVG
MAV_3743|M.avium_104 MANFTAADVKRLRELTGAGMLDCKNALAESDGDFDKAVEALRIKGAKDVG
Mb2913c|M.bovis_AF2122/97 MANFTAADVKRLRELTGAGMLACKNALAETDGDFDKAVEALRIKGAKDVG
Rv2889c|M.tuberculosis_H37Rv MANFTAADVKRLRELTGAGMLACKNALAETDGDFDKAVEALRIKGAKDVG
MLBr_01597|M.leprae_Br4923 MANFTVADVKRLRALTGAGMLDCKSVLVETDGNFDKAVESLRIKGAKDVG
Mflv_4132|M.gilvum_PYR-GCK MANYTAADVKRLRELTGAGMMDSKNALVEAEGDFDKAVELLRIKGAKDVG
Mvan_2210|M.vanbaalenii_PYR-1 MANYTAADVKRLRELTGAGMLDSKNALVEAEGDFDKAVELLRIKGAKDVG
MSMEG_2520|M.smegmatis_MC2_155 MANYTAADVKRLRELTGAGMLDSKNALVEADGDFDKAVELLRIKGAKDVG
TH_0149|M.thermoresistible__bu MANYTAADVKRLRELTGAGMLDSKNALVEAEGDFDKAVELLRIKGAKDVG
MAB_3195c|M.abscessus_ATCC_199 MANFTAADVKRLRELTGAGMLDCKNALAESDGDFDKAVEVLRIKGAKDVG
***:*.******* ******: .*..*.*::*:****** **********
MMAR_1820|M.marinum_M KRAERATAEGLVAAKDGALIELNCETDFVAKNAEFQKLADDIVAAAVASK
MUL_2068|M.ulcerans_Agy99 KRAERATAEGLVAAKDGALIELNCETDFVAKNAEFQKLADDIVAAAVASK
MAV_3743|M.avium_104 KRAERATAEGLVAAQGGALIELNSETDFVAKNAEFQALADQIVAAAASSK
Mb2913c|M.bovis_AF2122/97 KRAERATAEGLVAAKDGALIELNCETDFVAKNAEFQTLADQVVAAAAAAK
Rv2889c|M.tuberculosis_H37Rv KRAERATAEGLVAAKDGALIELNCETDFVAKNAEFQTLADQVVAAAAAAK
MLBr_01597|M.leprae_Br4923 KRAERATAEGLVAAKDGALIELNCETDFVAKNAEFQKLANQIVGVVAAAK
Mflv_4132|M.gilvum_PYR-GCK KRAERATAEGLVAAKDGALIELNSETDFVAKNAEFQAVAEQIVAAAAAAK
Mvan_2210|M.vanbaalenii_PYR-1 KRAERATAEGLVAAKDGALIELNSETDFVAKNGEFQALADQIVAAAAAAK
MSMEG_2520|M.smegmatis_MC2_155 KRAERATAEGLVAAKDGALIELNSETDFVAKNAEFQALADQIVAAAVAAK
TH_0149|M.thermoresistible__bu KRAGRATAEGLVAAKDGALIELNSETDFVAKNEEFQQLADAVVAAAAAAK
MAB_3195c|M.abscessus_ATCC_199 KRAERATAEGLVVARDGALVELNSETDFVAKNAEFQALADAIVGAAAAGK
*** ********.*:.***:***.******** *** :*: :*....:.*
MMAR_1820|M.marinum_M AADVDALKAASIGG-QTVEEAIGALSAKIGEKLELRRVAIFGGTVETYLH
MUL_2068|M.ulcerans_Agy99 AADVDALKAASIGG-QTVEEAIGALSAKIGEKLELRRVAIFGGTVETYLH
MAV_3743|M.avium_104 AADVDALKAAKIGD-TTVEQAIAELSAKIGEKLELRRVAHFDGTVEAYLH
Mb2913c|M.bovis_AF2122/97 PADVDALKGASIGD-KTVEQAIAELSAKIGEKLELRRVAIFDGTVEAYLH
Rv2889c|M.tuberculosis_H37Rv PADVDALKGASIGD-KTVEQAIAELSAKIGEKLELRRVAIFDGTVEAYLH
MLBr_01597|M.leprae_Br4923 IVDVDALKGASVGD-KTVEQAIAELAAKIGEKLKLRRAAIFNGTVATYLH
Mflv_4132|M.gilvum_PYR-GCK ATDVDALKAAKLGD-TTVEQTIADLSAKIGEKLELRRATYFDGQVETYLH
Mvan_2210|M.vanbaalenii_PYR-1 AADVDALKAVKLGD-TTVEQAIADLSAKIGEKLELRRATYFDGTTETYLH
MSMEG_2520|M.smegmatis_MC2_155 ANDIETLKAAKTGD-TTVEQAIADLSAKIGEKLELRRAAYFDGTVEAYLH
TH_0149|M.thermoresistible__bu ATDVDTLKAAKVGD-KTVEEAIAELAAKIGEKLELRRVAYFDGTVSTYLH
MAB_3195c|M.abscessus_ATCC_199 IGEVDALLAAKVEDGRTVEQAIADLAAKIGEKLELRRVAYFGGTVEAYLH
::::* ... . ***::*. *:*******:***.: *.* . :***
MMAR_1820|M.marinum_M RRAADLPPAVGVLVEY-TGAG----AEAAHAVALQIAALKARYLSREDVP
MUL_2068|M.ulcerans_Agy99 RRAADLPPAVGVLVEY-TGAG----AEAAHAVALQIAALKARYLSREDVP
MAV_3743|M.avium_104 RRAADLPPAVGVLVEY-QGSGKDSDKEAAHAVALQIAALKARYLSRDDVP
Mb2913c|M.bovis_AF2122/97 RRSADLPPAVGVLVEY-RGDD----AAAAHAVALQIAALRARYLSRDDVP
Rv2889c|M.tuberculosis_H37Rv RRSADLPPAVGVLVEY-RGDD----AAAAHAVALQIAALRARYLSRDDVP
MLBr_01597|M.leprae_Br4923 KRAADLPPAVGVLVEYGAGTDAANSTAAAHAAALQIAALKARFLSRDDVP
Mflv_4132|M.gilvum_PYR-GCK KRAADLPPAVGVLVEYT-GDDKS----AAHAVALQIAALKAKYLTREDVP
Mvan_2210|M.vanbaalenii_PYR-1 KRAADLPPAVGVLVEYT-GGDTS----AAHAVALQIAALKAKYLTREDVP
MSMEG_2520|M.smegmatis_MC2_155 KRAADLPPAVGVLVEYQ-AGDADKGKEAAHAVALQIAALKAKYLTREDVP
TH_0149|M.thermoresistible__bu KRAADLPPAVGVMVEYT-AEDAAKGAEAAHAVALQIAAMKAQYLTRDDVP
MAB_3195c|M.abscessus_ATCC_199 KRAADLPPAVGVLVEYESASGDATGADAAHAVALQIAALKAKYLTREDVP
:*:*********:*** . . ****.******::*::*:*:***
MMAR_1820|M.marinum_M EDLVASERRIAEETAKEEGKPEQALPKIVEGRLNGFFKDAVLLEQPSVSD
MUL_2068|M.ulcerans_Agy99 EDLVASERRIAEETAKEEGKPEQALPKIVEGRLNGFFKDAVLLEQPSVSD
MAV_3743|M.avium_104 EDVVASERRIAEETAKAEGKPEQALPKIVEGRLNGFFKDAVLLEQPSVSD
Mb2913c|M.bovis_AF2122/97 EDIVASERRIAEETARAEGKPEQALPKIVEGRLNGFFKDAVLLEQASVSD
Rv2889c|M.tuberculosis_H37Rv EDIVASERRIAEETARAEGKPEQALPKIVEGRLNGFFKDAVLLEQASVSD
MLBr_01597|M.leprae_Br4923 EDVLASERRIAEETAKAAGKPEQSLPKIVEGRLNGFFKDSVLLEQPSVFD
Mflv_4132|M.gilvum_PYR-GCK EDIVANERRIAEETARAEGKPEQALTKIVEGRVTGFYKDVVLLDQPSVSD
Mvan_2210|M.vanbaalenii_PYR-1 ADIVANERRIAEETARNEGKPEQALSKIVEGRVTGFYKDVVLLDQPAVSD
MSMEG_2520|M.smegmatis_MC2_155 EDIVANERRIAEETARNEGKPEQALPKIVEGRVTGFYKDVVLLDQPSVSD
TH_0149|M.thermoresistible__bu EDVVANERRIAEETAKAEGKPEQALPKIVEGRLNGFFKDVVLLDQPTVSD
MAB_3195c|M.abscessus_ATCC_199 ADVVENERRIAEETAKAEGKPEAALPKIVEGRVTGYYKDVVLLDQPSVSD
*:: .*********: **** :*.******:.*::** ***:*.:* *
MMAR_1820|M.marinum_M SKKSVKALLDDAGVTVTQFVRFEVGQA
MUL_2068|M.ulcerans_Agy99 SKKSVKALLDDAGVTVTQFVRFEVGQA
MAV_3743|M.avium_104 SKKTVKALLDEAGVTVTRFVRFEVGQA
Mb2913c|M.bovis_AF2122/97 NKKTVKALLDVAGVMVTRFVRFEVGQA
Rv2889c|M.tuberculosis_H37Rv NKKTVKALLDVAGVTVTRFVRFEVGQA
MLBr_01597|M.leprae_Br4923 NKKTVKVLLDEAGVTVTRFVRFEVGQA
Mflv_4132|M.gilvum_PYR-GCK NKKSVKALLDEAGVTVTRFARFEVGQA
Mvan_2210|M.vanbaalenii_PYR-1 NKKSVKALLDEAGVTVTRFVRFEVGQA
MSMEG_2520|M.smegmatis_MC2_155 NKKTVKALLDEAGVTVTRFVRFEVGQA
TH_0149|M.thermoresistible__bu NKKTVKALLDEAGVTVTRFVRFEVGQA
MAB_3195c|M.abscessus_ATCC_199 NKKSVKALLDEAGVTVTRFARFEVGQA
.**:**.*** *** **:*.*******