For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TMFEVDGLGSAERAGSGVRVSAEVVHSYDLTVIAADVDGVVASAGGWLCDRVRAGWQVTVLVAAGADTGA LTVLGVRDETLDSAAAALRRPTAAVAVDTRVLTHDDNARTEVLRLLDSGRTEVTVWGTPSPVTSELLSDA RFDTVCHRASAAARAFKAGALRTCGHDATGPSVETFISAGLWYPPDGGDLVPVSGT*
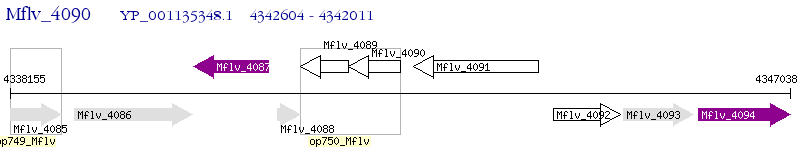
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4090 | - | - | 100% (197) | hypothetical protein Mflv_4090 |
| M. gilvum PYR-GCK | Mflv_2448 | - | 5e-25 | 46.51% (172) | hypothetical protein Mflv_2448 |
| M. gilvum PYR-GCK | Mflv_4089 | - | 4e-19 | 36.36% (165) | hypothetical protein Mflv_4089 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4738 | - | 8e-20 | 44.85% (136) | hypothetical protein MMAR_4738 |
| M. avium 104 | MAV_5125 | - | 2e-20 | 43.17% (139) | hypothetical protein MAV_5125 |
| M. smegmatis MC2 155 | MSMEG_4830 | - | 4e-21 | 41.04% (173) | hypothetical protein MSMEG_4830 |
| M. thermoresistible (build 8) | TH_0257 | - | 1e-22 | 44.25% (174) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1047 | - | 3e-16 | 40.00% (125) | hypothetical protein MUL_1047 |
| M. vanbaalenii PYR-1 | Mvan_2254 | - | 7e-53 | 55.90% (195) | hypothetical protein Mvan_2254 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4830|M.smegmatis_MC2_155 --------------------------MRYRLDVVAPCVIDAVRYAGGWLV
TH_0257|M.thermoresistible__bu --------------------------MRYRLDVVAHDVVDVVRNAGGWIV
MMAR_4738|M.marinum_M --------------------------MRYRLDVVASTVIDVVRYAGGWLF
MAV_5125|M.avium_104 ---MVAPTFDISKSSAARVTRANDEGLRYRLDVVAASAVDVVQSAGGWLY
MUL_1047|M.ulcerans_Agy99 ---------------------------------MAASVIDVVQAAGGWLC
Mflv_4090|M.gilvum_PYR-GCK MTMFEVDGLGSAERAGSGVRVSAEVVHSYDLTVIAADVDGVVASAGGWLC
Mvan_2254|M.vanbaalenii_PYR-1 --MFGVDDLAHFDRKGSGVRISGETLHTYDLRVVAGDVGGVVAAAGGWLC
:* . ..* ****:
MSMEG_4830|M.smegmatis_MC2_155 DRVMAGWDVTVLIDRDEDVRPLEILGVEAMDLESVLRSWQDRPHPQTVAV
TH_0257|M.thermoresistible__bu DRVMAGWDVTVLTEDRADIRPLQILGVATLPLEDVLASWEGRPHPQTVAV
MMAR_4738|M.marinum_M DRAMAGWDVTVLVADHLDDRPLHILGAQTLDLECALASVATRPRPQALAA
MAV_5125|M.avium_104 DRAMAGWEVTVLLPHGCNTRSLRILGVRAADLDSGLAGLG--QGAAGLAV
MUL_1047|M.ulcerans_Agy99 DRVMAGWQVRVLVPGGDDIRPLQILGATAMDLEAGLTGRA--PMGHSLAV
Mflv_4090|M.gilvum_PYR-GCK DRVRAGWQVTVLVAAGADTGALTVLGVRDETLDS-AAAALRR-PTAAVAV
Mvan_2254|M.vanbaalenii_PYR-1 DRVRAGWKVTVLVPPGHDVRALTILGVGVDAGES-AAEEPGLDSAAAIAV
**. ***.* ** : .* :**. : :*.
MSMEG_4830|M.smegmatis_MC2_155 AADLLNTDKRVRHGVLGALEQGLTEVTLWGD----ECPADLR--VCPAHH
TH_0257|M.thermoresistible__bu AADLFDRDPRVRRGVLGALEQGHTEVTLWGD----RWPAELSDGIGAVQH
MMAR_4738|M.marinum_M GADLFGCDPRVRRGVLQAIDHGVTEVTLWGE----SWPAELEASVSRVLH
MAV_5125|M.avium_104 SADAFTADARVRDTVLESLHNRLTAVALWGD----GWPLGVSRATTRAQH
MUL_1047|M.ulcerans_Agy99 GAAAFAADERVRAKLTEALQCRWTEVALWGQ----GWPLGVDRAIAPVQH
Mflv_4090|M.gilvum_PYR-GCK DTRVLTHDDNARTEVLRLLDSGRTEVTVWGTPSPVTSELLSDARFDTVCH
Mvan_2254|M.vanbaalenii_PYR-1 DARVLSGDEQLREDVLRLVDAARAEITVWGG----SALFGSDGRFGRVRH
: : * . * : :. : :::** . *
MSMEG_4830|M.smegmatis_MC2_155 ELSAAARAFKAQALTAANHPDAGSVGSTEAFRTGTMLSRSVPADLITAS-
TH_0257|M.thermoresistible__bu QLSAAARAFKAQALAAANDLEADFVTGTETFRSGVMTRLPMVSDLIPAS-
MMAR_4738|M.marinum_M PLSAAARTFKAAALAAA-AAAPGAVDGSEIFRSGLVARDSVAADLVPA--
MAV_5125|M.avium_104 LLSGAARRFKGYALAAEGISDGGIAPAESLLCDVCSAADSELVRLD----
MUL_1047|M.ulcerans_Agy99 VLSAAALAFKR---------------------------------------
Mflv_4090|M.gilvum_PYR-GCK RASAAARAFKAGALRTCGHDATGPS--VETFISAGLWYPPDGGDLVPVSG
Mvan_2254|M.vanbaalenii_PYR-1 RLSAAARAFKARALEATGQPTPDLA--VEEFVSAALWYPTDGADLVPHQ-
*.** **
MSMEG_4830|M.smegmatis_MC2_155 -
TH_0257|M.thermoresistible__bu -
MMAR_4738|M.marinum_M -
MAV_5125|M.avium_104 -
MUL_1047|M.ulcerans_Agy99 -
Mflv_4090|M.gilvum_PYR-GCK T
Mvan_2254|M.vanbaalenii_PYR-1 -