For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RATGLISEFGVDASLFVSHGLCTPHGILRNAQFGRVFRRPGVRTVLSRGCARSQSASLGFPAASKVPRGP LRLDDAGWRPRNRLVAWGFRVGICRSATREPTPHAYGPVFKRHLPGYPSAGRGPSEEQNMIGTIIGAIVV GLIVGALARLVMPGKQNMGVLMTIVLGALGSFLGTWITYTLGYSNSNGGFEVIPFLVGIIVAIALIAAYL GITGRRGTRPRV*
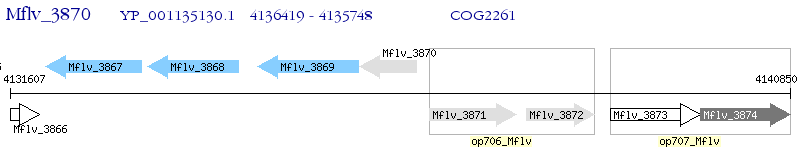
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3870 | - | - | 100% (223) | membrane protein-like protein |
| M. gilvum PYR-GCK | Mflv_3765 | - | 8e-15 | 43.21% (81) | hypothetical protein Mflv_3765 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2437c | - | 3e-09 | 39.13% (92) | hypothetical protein MAB_2437c |
| M. marinum M | MMAR_2567 | - | 2e-14 | 40.96% (83) | hypothetical protein MMAR_2567 |
| M. avium 104 | MAV_2858 | - | 2e-05 | 32.88% (73) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_3022 | - | 8e-14 | 38.89% (90) | transglycosylase associated protein |
| M. thermoresistible (build 8) | TH_4623 | - | 5e-23 | 51.72% (87) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_3194 | - | 1e-14 | 40.96% (83) | hypothetical protein MUL_3194 |
| M. vanbaalenii PYR-1 | Mvan_2638 | - | 3e-14 | 41.11% (90) | putative transglycosylase associated protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3870|M.gilvum_PYR-GCK MRATGLISEFGVDASLFVSHGLCTPHGILRNAQFGRVFRRPGVRTVLSRG
TH_4623|M.thermoresistible__bu --------------------------------------------------
MMAR_2567|M.marinum_M --------------------------------------------------
MUL_3194|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3022|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_2638|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2437c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2858|M.avium_104 --------------------------------------------------
Mflv_3870|M.gilvum_PYR-GCK CARSQSASLGFPAASKVPRGPLRLDDAGWRPRNRLVAWGFRVGICRSATR
TH_4623|M.thermoresistible__bu --------------------------------------------------
MMAR_2567|M.marinum_M --------------------------------------------------
MUL_3194|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3022|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_2638|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2437c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2858|M.avium_104 -----------------------------------------------MTA
Mflv_3870|M.gilvum_PYR-GCK EPTPHAYGPVFKRHLPGYPSAGRGPSEEQNMIGTIIGAIVVGLIVGALAR
TH_4623|M.thermoresistible__bu -------------------------------MGTIIVALIVGALLGALAR
MMAR_2567|M.marinum_M -----------------------------MTITGVITAILIGTVIGVLGR
MUL_3194|M.ulcerans_Agy99 -----------------------------MTITGVITAILIGTVIGVLGR
MSMEG_3022|M.smegmatis_MC2_155 -----------------------------MTITGIISAILIGIVVGVIGR
Mvan_2638|M.vanbaalenii_PYR-1 -----------------------------MTITGIISAILIGIVVGVLGR
MAB_2437c|M.abscessus_ATCC_199 ------------------------------MVLNIIWMIILGLIVGWIAR
MAV_2858|M.avium_104 QQTAGLCQRLRGDPHGRHCCHRIPARSTTLTSVGIIGYIIIGGLAGALAS
:* :::* : * :.
Mflv_3870|M.gilvum_PYR-GCK LVMPGKQNMGVLMTIVLGALGSFLGTWITYTLGYSNSNGGFEVI--PFLV
TH_4623|M.thermoresistible__bu LILPGKQNIGMLITVLLGAVGALLGSWISNAM-FGTGDKMFSPI--PFVI
MMAR_2567|M.marinum_M LVVPGRQSIGVLVTILVGIVSAFIGTAIAKAVGIPTATSGVDWL--ELLV
MUL_3194|M.ulcerans_Agy99 LVVPGRQSIGVLVTILVGIVSAFIGTAIAKAVGIPTATSGVDWL--ELLV
MSMEG_3022|M.smegmatis_MC2_155 LIVPGKQPIGFLVTILVGIVAAFIGTALARAMGIPTATSGIDWL--ELLV
Mvan_2638|M.vanbaalenii_PYR-1 LALPGKQPIGMLVTILVGIVSAFLGTALARVIGIPTATSGVDWL--ELLV
MAB_2437c|M.abscessus_ATCC_199 LIVPGKENFGWIATALLGIIGGYVGGTLGSLVFAPHKFTVTPPINHAFLG
MAV_2858|M.avium_104 KIVRG-SGAGILMDIVIGIVGALIGGFILSFFVNTAGGG--------LIF
: * . * : ::* :.. :* : . ::
Mflv_3870|M.gilvum_PYR-GCK GIIVAIALIAAYLGITGRRGTRPRV--------
TH_4623|M.thermoresistible__bu GLVVAVLLIAIYAGVAGRRG-APRA--------
MMAR_2567|M.marinum_M QVIVAAVGVAAVASLMGRRHSGITGGRRSGLLH
MUL_3194|M.ulcerans_Agy99 QVIVAAVGVAAVASLMGRRHSGITGGRRSGLLH
MSMEG_3022|M.smegmatis_MC2_155 QVVVAAIGVALVAAVMG-RRTGVLGRRRSGLMR
Mvan_2638|M.vanbaalenii_PYR-1 QVVVAAIGVALVAALMGRRRTGVTGRG-SGLTR
MAB_2437c|M.abscessus_ATCC_199 ALVGAVILLVAYKFIAGKTKS------------
MAV_2858|M.avium_104 TFFTALLGSVILLWIVGMVRRT-----------
.. * . : *