For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GDVAAGVAAAGVLVWLIALSAFDIRHRRLPNVLTLPGAAVILVAATVVGHGGPALLGATALFAVYVTVHL FSPGALGAGDVKLAIGLGALTGAYGADVWVVAALGASLMTGALALCRRLQGAGPDVPHGPSMCLAAGTAL GLAALA*
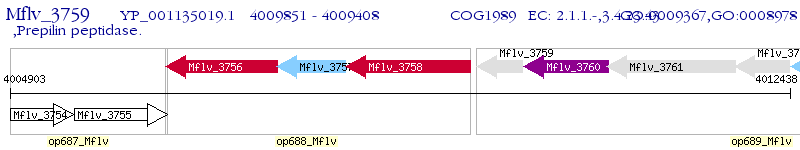
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3759 | - | - | 100% (147) | peptidase A24A, prepilin type IV |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2581c | - | 7e-36 | 56.20% (137) | hypothetical protein Mb2581c |
| M. tuberculosis H37Rv | Rv2551c | - | 7e-36 | 56.20% (137) | hypothetical protein Rv2551c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2844c | - | 1e-34 | 54.35% (138) | hypothetical protein MAB_2844c |
| M. marinum M | MMAR_2174 | - | 1e-39 | 60.29% (136) | hypothetical protein MMAR_2174 |
| M. avium 104 | MAV_3427 | - | 2e-40 | 58.70% (138) | peptidase, A24 (type IV prepilin peptidase) family protein |
| M. smegmatis MC2 155 | MSMEG_3029 | - | 3e-43 | 61.27% (142) | peptidase, A24 (type IV prepilin peptidase) family protein |
| M. thermoresistible (build 8) | TH_3696 | - | 2e-41 | 57.75% (142) | PUTATIVE oligopeptide/dipeptide ABC transporter, ATPase subunit |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2644 | - | 3e-52 | 67.36% (144) | peptidase A24A, prepilin type IV |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2844c|M.abscessus_ATCC_199 ------MIYGAGVAVLCWMTALSYFDIRYRRLPNWLTLPAAVVIIT-VGL
MSMEG_3029|M.smegmatis_MC2_155 MGLSSGALGAVAVVAAAWLIALSGYDLRLRRLPNWLTVPGAVLILVGATL
Mflv_3759|M.gilvum_PYR-GCK --MGDVAAGVAAAGVLVWLIALSAFDIRHRRLPNVLTLPGAAVILVAATV
Mvan_2644|M.vanbaalenii_PYR-1 --MGRAGAAVAVIGVSGWLIALCAFDIRQRRLPNMLTVPGAAVILAVAAL
Mb2581c|M.bovis_AF2122/97 ---------MLAAAVLAWMGVLCVCDVRQRRLPNWLTLPGAGVILLFAGL
Rv2551c|M.tuberculosis_H37Rv ---------MLAAAVLAWMGVLCVCDVRQRRLPNWLTLPGAGVILLFAGL
TH_3696|M.thermoresistible__bu ------MGWAGAVVALGWLATLSVCDIRWRRLPNRLTLPGAVVVLMAAAA
MMAR_2174|M.marinum_M --MWLVIRTPVVAVVVAWLLVLSYTDIRHRRLPNRFTLPGAGAMLLGALV
MAV_3427|M.avium_104 ------MRIAAAGAVLTWLAVLSCYDIRQHRLPNALTLTGAAAILAAAGL
. *: .*. *:* :**** :*:..* :: .
MAB_2844c|M.abscessus_ATCC_199 WGRGPSIWAGAAALTALYLAAHLLSPRAMGAGDVKLAVSTGGLAGAFGLD
MSMEG_3029|M.smegmatis_MC2_155 AGRGAAALAGAAALSALYLVVHLVSPRSLGGGDVKLAVGLGALTGMFGAD
Mflv_3759|M.gilvum_PYR-GCK VGHGGPALLGATALFAVYVTVHLFSPGALGAGDVKLAIGLGALTGAYGAD
Mvan_2644|M.vanbaalenii_PYR-1 AGHGRPAALGAAALFTVYATVHLLAPAAMGAGDVKLAIGLGALTGVFGVD
Mb2581c|M.bovis_AF2122/97 AGRGVPALAGAAALAGVYLLVHLALPAAMGAGDVKLAIGLGGLTGCFGVE
Rv2551c|M.tuberculosis_H37Rv AGRGVPALAGAAALAGVYLLVHLALPAAMGAGDVKLAIGLGGLTGCFGVE
TH_3696|M.thermoresistible__bu AGHGVPALLGSMALTGVYLIVHLLAPAAMGAGDVKLALGLGGMTGAFGPE
MMAR_2174|M.marinum_M AGRGTAALAGAAALTSVYLLVHLLAPAALGAGDVKLAIGLGALAGSLGVE
MAV_3427|M.avium_104 AGRGPSALAGAAALAAIYLLVHAVAPGGMGAGDVKLALGVGALTGCGGVG
*:* . *: ** :* .* * .:*.******:. *.::* *
MAB_2844c|M.abscessus_ATCC_199 AWFLAAIGAPLVTGLTGLVLLGLGRRGAT-VPHGPSMCLATAVAAGLGAT
MSMEG_3029|M.smegmatis_MC2_155 VWWLAALGAPLLT---GVVALGAALRGATTVPHGPSMCVASAVAVGLVLL
Mflv_3759|M.gilvum_PYR-GCK VWVVAALGASLMTGALALCRRLQ--GAGPDVPHGPSMCLAAGTALGLAAL
Mvan_2644|M.vanbaalenii_PYR-1 AWVLAAVGASVLTGVLALCLRMA--GAGATVPHGPSMCLSTAAVLALALP
Mb2581c|M.bovis_AF2122/97 VWFLAALAAPLLTAVCGVMVTP---WGVRTLPHGPSMCVASLGAVGLALL
Rv2551c|M.tuberculosis_H37Rv VWFLAALAAPLLTAVCGVMVTP---WGVRTLPHGPSMCVASLGAVGLALL
TH_3696|M.thermoresistible__bu VWTLAALAAPLLTGVLGVIALIR--SAARTVPHGPSMCVASAGAVALAIL
MMAR_2174|M.marinum_M VWFLAALAAPLMTALWAVGARLR--TGALAVPHGPSMCLATAVAAGLALL
MAV_3427|M.avium_104 VWFLAALAAPLLTALVGVLARVR--RAGPAVPHGPSLCLAAAAGAGLALF
.* :**:.*.::* .: . :*****:*::: .*
MAB_2844c|M.abscessus_ATCC_199 TPGI
MSMEG_3029|M.smegmatis_MC2_155 ----
Mflv_3759|M.gilvum_PYR-GCK A---
Mvan_2644|M.vanbaalenii_PYR-1 TQGV
Mb2581c|M.bovis_AF2122/97 G---
Rv2551c|M.tuberculosis_H37Rv G---
TH_3696|M.thermoresistible__bu ----
MMAR_2174|M.marinum_M G---
MAV_3427|M.avium_104 ----