For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IRIAQLLLILAAAGLWAASRMTWVQISSFDGLGQPKTTDLSGSTWSTALVPLALLILAAAVAALAVRGWP LRILAVLVAAASAGMGYLAISLWVVADVAVRAARLAEVPVADLLDTQRDYVGAVITLIAAALSLVGAVVM MRSAARGRAETARYTRRPAAEAPTEASAMSERMIWDALDEGRDPTNPDNKGR*
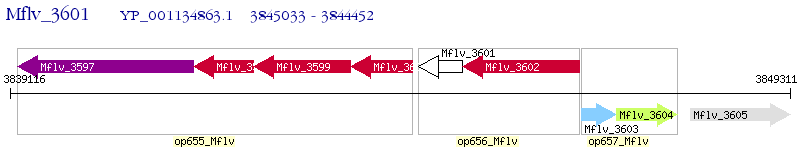
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3601 | - | - | 100% (193) | hypothetical protein Mflv_3601 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1636 | - | 2e-54 | 55.71% (219) | hypothetical protein Mb1636 |
| M. tuberculosis H37Rv | Rv1610 | - | 2e-54 | 55.71% (219) | hypothetical protein Rv1610 |
| M. leprae Br4923 | MLBr_01270 | - | 6e-53 | 54.37% (206) | hypothetical protein MLBr_01270 |
| M. abscessus ATCC 19977 | MAB_2646c | - | 2e-44 | 49.18% (183) | hypothetical protein MAB_2646c |
| M. marinum M | MMAR_2412 | - | 4e-52 | 53.02% (215) | hypothetical protein MMAR_2412 |
| M. avium 104 | MAV_3176 | - | 3e-57 | 56.81% (213) | hypothetical protein MAV_3176 |
| M. smegmatis MC2 155 | MSMEG_3218 | - | 9e-66 | 69.84% (189) | hypothetical protein MSMEG_3218 |
| M. thermoresistible (build 8) | TH_3421 | - | 1e-66 | 64.97% (197) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_1590 | - | 4e-51 | 52.56% (215) | hypothetical protein MUL_1590 |
| M. vanbaalenii PYR-1 | Mvan_2815 | - | 6e-86 | 81.35% (193) | hypothetical protein Mvan_2815 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3601|M.gilvum_PYR-GCK ----------------MIRIAQLLLILAAAGLWAASRMTWVQISSFDGLG
Mvan_2815|M.vanbaalenii_PYR-1 ----------------MTRVAQLLLVLAAAGLWAASRLTWVQITSFDGLG
MSMEG_3218|M.smegmatis_MC2_155 ----------------MTRAAQALLVVAALGLWGASRFNWVEVRSFDGLG
TH_3421|M.thermoresistible__bu ----------------VIRVGQVLLVLAAVGLWVASRMTWVQVTTFDGLT
MMAR_2412|M.marinum_M ----------------MIGIAQLLLVISAGVLWIASRLGWVVIRSFDGLG
MUL_1590|M.ulcerans_Agy99 MTDTGNDRRARRNSRLMIGIAQLLLVISAGVLWIASRLGWVVIRSFDGLG
Mb1636|M.bovis_AF2122/97 MAANAGSVRPNRRARPMIGIAQLLLVVAAGALWMAARLPWVVIGSFDELG
Rv1610|M.tuberculosis_H37Rv MAANAGSVRPNRRARPMIGIAQLLLVVAAGALWMAARLPWVVIGSFDELG
MLBr_01270|M.leprae_Br4923 --MMAPDIKSARAGRLTIQIAQLLLVVAAGALWMAARLPWVVIRSFDGLG
MAV_3176|M.avium_104 ---MTEARARRLDGRWLIRIAQLLLVIAAVGLWVAARLPWVVIRSFDGLG
MAB_2646c|M.abscessus_ATCC_199 ----MSDPAPRNPGRRRLLMGAGLLVIAAGALWVASKFSWVRLRSFDGLG
. **:::* ** *::: ** : :** *
Mflv_3601|M.gilvum_PYR-GCK QPKTTDLSGSTWSTALVPLALLILAAAVAALAVRGWPLRILAVLVAAASA
Mvan_2815|M.vanbaalenii_PYR-1 QPKTADLSGATWSTALIPLALLVLAAAGAALAVRGWPLRVLAVLVAAASA
MSMEG_3218|M.smegmatis_MC2_155 QPKTTALTGASWSTALIPLALLALAAAVAALAVRGWMLRLLAILVAAASA
TH_3421|M.thermoresistible__bu HPKTTPVDGVTWSTALVPLALLSLAAAVAALAVRGWALRLLALLVALAGA
MMAR_2412|M.marinum_M PPKEVTLSGATWSTALLPMALLMLAAAVAALAVRGWPLRGLAALLAMVSL
MUL_1590|M.ulcerans_Agy99 PPKEVTLSGATWSTALLPMALLMLAAAVAALAVRGWPLRGLAALLAMVSL
Mb1636|M.bovis_AF2122/97 PPKEVTLTGASWSTALLPLALLMLAAAVAALAVRGWPLRALAVLLAAASF
Rv1610|M.tuberculosis_H37Rv PPKEVTLTGASWSTALLPLALLMLAAAVAALAVRGWPLRALAVLLAAASF
MLBr_01270|M.leprae_Br4923 PPKEVALSGASWSAVLLPLALLMLAATVAAIAVRGWPLRVLAGLLAVASF
MAV_3176|M.avium_104 PPKAVTLSGGTWSTALLPLALLLLAAAVAAIAVRGWLLRALAGLLAVASL
MAB_2646c|M.abscessus_ATCC_199 EPRTLLVSGADWSAALVPLAVALIAAAIATLAVRGWALRVLAVLVAIIGA
*: : * **:.*:*:*: :**: *::***** ** ** *:* .
Mflv_3601|M.gilvum_PYR-GCK GMGYLAISLWVVADVAVRAARLAEVPVADLLDTQRDYVGAVITLIAAALS
Mvan_2815|M.vanbaalenii_PYR-1 GMGYLAISLWVMADDAVRAARLADVPVADLLDTQRHYGGAGVTLVAGVLS
MSMEG_3218|M.smegmatis_MC2_155 GMGYLAISLWAVRDVAVRAADLALVPVAQLTGTHRYYTGAVITLAAAVCT
TH_3421|M.thermoresistible__bu GMAYLGISLWVVPDVAVRAVHLVELSVIDLVDSQRLYGGAVVTVASAVGA
MMAR_2412|M.marinum_M AVGYLGVSLWVIPDVALRGAELAQVSVITLVGSGRHYVGAGLAVGAAVCT
MUL_1590|M.ulcerans_Agy99 AVGYLGVSLWVIPDVALRGAELAQVSVITLVGSGRHYVGAGLAVGAVVCT
Mb1636|M.bovis_AF2122/97 AVGYLGISLWVVPDVAARGADLAHVPVVTLVGSARHYWGAVAAVLAAVCA
Rv1610|M.tuberculosis_H37Rv AVGYLGISLWVVPDVAARGADLAHVPVVTLVGSARHYWGAVAAVLAAVCA
MLBr_01270|M.leprae_Br4923 LVGYLGVSLWVLPDVTVRGAVLAHVSLLSLVGSQRHHLGAGAAVAASGCT
MAV_3176|M.avium_104 AVGYLGVSLWAVPDVAVRGTELAHLSVISLVGSERRYLGAGITVAAAALT
MAB_2646c|M.abscessus_ATCC_199 AAAYLGVSLWAVRDVGPRAAEVKNVAVSTLVGADRLSGGAITAVLAAVVS
.**.:***.: * *.. : :.: * .: * ** :: : :
Mflv_3601|M.gilvum_PYR-GCK LVGAVVMMRSAARG---RAETARYT----RRPAAEAPTEASAMS------
Mvan_2815|M.vanbaalenii_PYR-1 LAGAVLLMRSATHR---RAEAARYT----RRTPAESDTRAAAMS------
MSMEG_3218|M.smegmatis_MC2_155 LVGAVLLMRSANGSQ--RAVAARYAAPAARRTAAQHDVSSESMS------
TH_3421|M.thermoresistible__bu LVAAVLLMRAAAKP---ASGAAKYAAPAARRAAARRDSSDSAMS------
MMAR_2412|M.marinum_M LAAAALLMRSASIVESLGDNATKYASPASRRSVARAQDAAIATEKASKSA
MUL_1590|M.ulcerans_Agy99 LAAAALLMRLASIVESLGDNATKYATPASRRSVARAQDAAIATEKASKSA
Mb1636|M.bovis_AF2122/97 LLAAVFLMSSAAIRGSAGEDMARYAAPRARRSIARRQHSNAAGRAAPQDD
Rv1610|M.tuberculosis_H37Rv LLAAVFLMSSAAIRGSAGEDMARYAAPRARRSIARRQHSNAAGRAAPQDD
MLBr_01270|M.leprae_Br4923 LIAAVLLMRSASVIGSARQGTSKYVVPAQRRSIARRDGAATAIS------
MAV_3176|M.avium_104 LIAAVLLMRSASDPKSARSSAMKYATPATRRSITRRNAADGAMLEQPETP
MAB_2646c|M.abscessus_ATCC_199 VLAAITLLRAAR-----GASASKYQTPAARRSEAVSP-VDKGQS------
: .* :: * :* **. : .
Mflv_3601|M.gilvum_PYR-GCK ---------ERMIWDALDEGRDPT----NPDNKGR---------------
Mvan_2815|M.vanbaalenii_PYR-1 ---------ERMIWDALDEGHDPT----NPDNQGR---------------
MSMEG_3218|M.smegmatis_MC2_155 ---------ERMIWDALDEGHDPTG---DQDDTDRRGGDR----------
TH_3421|M.thermoresistible__bu ---------ERTMWDALDEGRDPT----DPDSMGR---------------
MMAR_2412|M.marinum_M ELDM----SERTIWDALDEGRDPTDRPPEPDTEGR---------------
MUL_1590|M.ulcerans_Agy99 ELDM----SERTIWDALDEGRDPTDRPPEPDTEGR---------------
Mb1636|M.bovis_AF2122/97 GPDMGPRMSERMIWEALDEGRDPTDREQESDTEGR---------------
Rv1610|M.tuberculosis_H37Rv GPDMGPRMSERMIWEALDEGRDPTDREQESDTEGR---------------
MLBr_01270|M.leprae_Br4923 ------QMSERMIWDALDEDRDPTDRLREPDTEGRWWTACRRSLPFMNVV
MAV_3176|M.avium_104 ------EMSERMIWDALDEGRDPTDRPSGSDTEGR---------------
MAB_2646c|M.abscessus_ATCC_199 ---------ERVMWDSLDDGHDPT---VGPAQT-----------------
** :*::**:.:***
Mflv_3601|M.gilvum_PYR-GCK -----------------------------
Mvan_2815|M.vanbaalenii_PYR-1 -----------------------------
MSMEG_3218|M.smegmatis_MC2_155 -----------------------------
TH_3421|M.thermoresistible__bu -----------------------------
MMAR_2412|M.marinum_M -----------------------------
MUL_1590|M.ulcerans_Agy99 -----------------------------
Mb1636|M.bovis_AF2122/97 -----------------------------
Rv1610|M.tuberculosis_H37Rv -----------------------------
MLBr_01270|M.leprae_Br4923 EIGGCTGSVAGRWVTSGKGNDTHVSGNCA
MAV_3176|M.avium_104 -----------------------------
MAB_2646c|M.abscessus_ATCC_199 -----------------------------