For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AFATEHPVLAHSEYRPVDDVVRSGARFEVVSEYEPAGDQPAAIEELERRIRAGEKDVVLLGATGTGKSAT TAWLIERLQRPTLVMAPNKTLAAQLANELREMLPNNAVEYFVSYYDYYQPEAYIAQTDTYIEKDSSINDD VERLRHSATSNLLSRRDVVVVASVSCIYGLGTPQSYMDRSVELKVGDEVPRDSLLRLLVDVQYTRNDMSF TRGTFRVRGDTVEIIPSYEELAVRIEFFGDEIEELYYLHPLTGDIIRKVDSLRIFPATHYVAGPERMAAA ISTIEAELEARLAELEGQGKLLEAQRLRMRTNYDIEMMRQVGFCSGIENYSRHIDGRPAGSAPATLLDYF PEDFLLVIDESHVTVPQIGGMYEGDMSRKRNLVDFGFRLPSAVDNRPLTWEEFADRIGQTVYLSATPGNF ELAQTGGEFVEQVIRPTGLVDPQVIVKPTKGQIDDLIGEIRKRTERDERVLVTTLTKKMAEDLTDYLLEM GIRVRYLHSEVDTLRRVELLRQLRLGEYDVLIGINLLREGLDLPEVSLVAILDADKEGFLRSPRSLIQTI GRAARNVSGEVHMYADKITDSMKQAIDETERRRAKQIAYNTERGIDPQPLRKKIADILDQVYREADDTEA VEIGGSGRNASRGRRAPGEPGRAVSAGVYEGRDTKNMPRAELADLIKDLTAQMMAAARDLQFELAARIRD EIQDLKKELRGMDAAGLK*
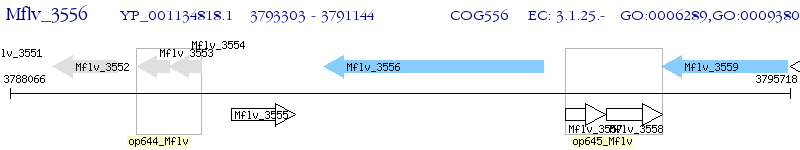
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3556 | - | - | 100% (719) | excinuclease ABC subunit B |
| M. gilvum PYR-GCK | Mflv_1948 | - | 6e-10 | 40.91% (88) | transcription-repair coupling factor |
| M. gilvum PYR-GCK | Mflv_2225 | - | 4e-06 | 22.36% (161) | DEAD/DEAH box helicase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1659 | uvrB | 0.0 | 91.12% (698) | excinuclease ABC subunit B |
| M. tuberculosis H37Rv | Rv1633 | uvrB | 0.0 | 91.12% (698) | excinuclease ABC subunit B |
| M. leprae Br4923 | MLBr_01387 | uvrB | 0.0 | 89.68% (698) | excinuclease ABC subunit B |
| M. abscessus ATCC 19977 | MAB_2308 | - | 0.0 | 89.57% (719) | excinuclease ABC subunit B |
| M. marinum M | MMAR_2437 | uvrB | 0.0 | 90.91% (726) | excinuclease ABC (subunit B-helicase) UvrB |
| M. avium 104 | MAV_3141 | uvrB | 0.0 | 91.70% (723) | excinuclease ABC subunit B |
| M. smegmatis MC2 155 | MSMEG_3816 | uvrB | 0.0 | 91.93% (719) | excinuclease ABC subunit B |
| M. thermoresistible (build 8) | TH_1003 | uvrB | 0.0 | 90.13% (719) | PROBABLE EXCINUCLEASE ABC (SUBUNIT B-HELICASE) UVRB |
| M. ulcerans Agy99 | MUL_1616 | uvrB | 0.0 | 91.14% (700) | excinuclease ABC subunit B |
| M. vanbaalenii PYR-1 | Mvan_3350 | - | 0.0 | 96.26% (722) | excinuclease ABC subunit B |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1659|M.bovis_AF2122/97 ------------------------------------------MRAGGHFE
Rv1633|M.tuberculosis_H37Rv ------------------------------------------MRAGGHFE
MLBr_01387|M.leprae_Br4923 ------------------------------------------MRVGGRFE
MAV_3141|M.avium_104 -----------------MAFATEHPVVAHSEYRPAEEAVEGLVRAGGRFE
MMAR_2437|M.marinum_M -----------------MAFATEHPADHVPVAHSEYRAVDDVVRTGASFE
MUL_1616|M.ulcerans_Agy99 ------------------------------------------MRTGASFE
Mflv_3556|M.gilvum_PYR-GCK -----------------MAFATEHPVLAHSEYR----PVDDVVRSGARFE
Mvan_3350|M.vanbaalenii_PYR-1 -----------------MAFATEHPVLAHSEYR----PVDEVVRSGARFE
MSMEG_3816|M.smegmatis_MC2_155 MGEHHRNLSVRSSTLVNMAFATEHPVLAHSEYR----AVDEIVRTGKPFE
TH_1003|M.thermoresistible__bu ---------------VNMAFATEHPVLAQSEYR----PVDAIVRTGGRFE
MAB_2308|M.abscessus_ATCC_1997 -----------------MAFATEHPIVAHSEYR----PVTDILRTEGRFE
:* **
Mb1659|M.bovis_AF2122/97 VVSPHAPAGDQPAAIDELERRINAGERDVVLLGATGTGKSATTAWLIERL
Rv1633|M.tuberculosis_H37Rv VVSPHAPAGDQPAAIDELERRINAGERDVVLLGATGTGKSATTAWLIERL
MLBr_01387|M.leprae_Br4923 VISPHEPAGDQPAAIDELQRRILAGERDVVLLGATGTGKSATTAWLIERL
MAV_3141|M.avium_104 VVSPHAPAGDQPAAIDELERRIRAGERDVVLLGATGTGKSATTAWLIERL
MMAR_2437|M.marinum_M VISPHDPAGDQPAAINELERRIRAGERDVVLLGATGTGKSATTAWLIERL
MUL_1616|M.ulcerans_Agy99 VISPHDPAGDQPAAINELERRIRAGERDVVLLGATGTGKSATTAWLIERL
Mflv_3556|M.gilvum_PYR-GCK VVSEYEPAGDQPAAIEELERRIRAGEKDVVLLGATGTGKSATTAWLIERL
Mvan_3350|M.vanbaalenii_PYR-1 VVSEFEPAGDQPAAIDELERRIRAGEKDVVLLGATGTGKSATTAWLIERL
MSMEG_3816|M.smegmatis_MC2_155 VVSPYAPAGDQPAAIDELERRIRAGERDVVLLGATGTGKSATTAWLIERL
TH_1003|M.thermoresistible__bu VISPYQPAGDQPAAIDELERRLRAGEKDVVLLGATGTGKSATTAWLIERM
MAB_2308|M.abscessus_ATCC_1997 VVSQYEPAGDQPGAIAELERRVRAGEKDVVLLGATGTGKSATTAWLIERL
*:* . ******.** **:**: ***:**********************:
Mb1659|M.bovis_AF2122/97 QRPTLVMAPNKTLAAQLANELREMLPHNAVEYFVSYYDYYQPEAYIAQTD
Rv1633|M.tuberculosis_H37Rv QRPTLVMAPNKTLAAQLANELREMLPHNAVEYFVSYYDYYQPEAYIAQTD
MLBr_01387|M.leprae_Br4923 QRPTLVMAPNKTLAAQLANELRGMLPHNAVEYFVSYYDYYQPEAYIAQTD
MAV_3141|M.avium_104 QRPTLVMAPNKTLAAQLANELREMLPHNAVEYFVSYYDYYQPEAYIAQTD
MMAR_2437|M.marinum_M QRPTLVMAPNKTLAAQLANELREMLPHNAVEYFVSYYDYYQPEAYIAQTD
MUL_1616|M.ulcerans_Agy99 QRPTLVMAPNKTLAAQLANELREMLPHNAVEYFVSYYDYYQPEAYIAQTD
Mflv_3556|M.gilvum_PYR-GCK QRPTLVMAPNKTLAAQLANELREMLPNNAVEYFVSYYDYYQPEAYIAQTD
Mvan_3350|M.vanbaalenii_PYR-1 QRPTLVMAPNKTLAAQLANELREMLPHNAVEYFVSYYDYYQPEAYIAQTD
MSMEG_3816|M.smegmatis_MC2_155 QRPTLVMAPNKTLAAQLANELREMLPHNAVEYFVSYYDYYQPEAYIAQTD
TH_1003|M.thermoresistible__bu QRPTLVIEPNKTLAAQMANELREMLPNNAVEYFVSYYDYYQPEAYIAQTD
MAB_2308|M.abscessus_ATCC_1997 QRPTLVMAPNKTLAAQLANELREMLPHNAVEYFVSYYDYYQPEAYIAQTD
******: ********:***** ***:***********************
Mb1659|M.bovis_AF2122/97 TYIEKDSSINDDVERLRHSATSALLSRRDVVVVASVSCIYGLGTPQSYLD
Rv1633|M.tuberculosis_H37Rv TYIEKDSSINDDVERLRHSATSALLSRRDVVVVASVSCIYGLGTPQSYLD
MLBr_01387|M.leprae_Br4923 TYIEKDSSINDDVERLRHSATSSLLSRRDVVVVASVSCIYGLGTPQSYLD
MAV_3141|M.avium_104 TYIEKDSSINDDVERLRHSATSSLLSRRDVVVVASVSCIYGLGTPQSYLD
MMAR_2437|M.marinum_M TYIEKDSSINDDVERLRHSATSALLSRRDVVVVASVSCIYGLGTPQSYLD
MUL_1616|M.ulcerans_Agy99 TYIEKDSSINDDVERLRHSATSALLSRRDVVVVASVSCIYGLGTPQSYLD
Mflv_3556|M.gilvum_PYR-GCK TYIEKDSSINDDVERLRHSATSNLLSRRDVVVVASVSCIYGLGTPQSYMD
Mvan_3350|M.vanbaalenii_PYR-1 TYIEKDSSINDDVERLRHSATSNLLSRRDVVVVASVSCIYGLGTPQSYMD
MSMEG_3816|M.smegmatis_MC2_155 TYIEKDSSINDDVERLRHSATSSLLSRRDVVVVASVSCIYGLGTPQSYLD
TH_1003|M.thermoresistible__bu TYIEKDSSINDDVERLRHSATSALLSRRDVVVVASVSCIYGLGTPQSYLD
MAB_2308|M.abscessus_ATCC_1997 TYIEKDSSINDDVERLRHSATSNLLSRRDVVVVASVSCIYGLGTPQSYLD
********************** *************************:*
Mb1659|M.bovis_AF2122/97 RSVELKVGEEVPRDGLLRLLVDVQYTRNDMSFTRGSFRVRGDTVEIIPSY
Rv1633|M.tuberculosis_H37Rv RSVELKVGEEVPRDGLLRLLVDVQYTRNDMSFTRGSFRVRGDTVEIIPSY
MLBr_01387|M.leprae_Br4923 RSVELAVSNEVPRDGLLRLLVDVQYTRNDLSFTRGSFRVRGDTVEIIPSY
MAV_3141|M.avium_104 RSVELRVGTEVPRDALLRLLVDVQYTRNDLSFTRGSFRVRGDTVEIIPSY
MMAR_2437|M.marinum_M RSVELQVGTDVPRDGLLRLLVDVQYTRNDLSFTRGSFRVRGDTVEIIPSY
MUL_1616|M.ulcerans_Agy99 RSVELQVGTDVPRDGLLRLLVDVQYTRNDLSFTRGSFRVRGDTVEIIPSY
Mflv_3556|M.gilvum_PYR-GCK RSVELKVGDEVPRDSLLRLLVDVQYTRNDMSFTRGTFRVRGDTVEIIPSY
Mvan_3350|M.vanbaalenii_PYR-1 RSVELKVGDEVPRDGLLRLLVDVQYTRNDMAFTRGTFRVRGDTVEIIPSY
MSMEG_3816|M.smegmatis_MC2_155 RSVELQVGQEVPRDGLLRLLVDVQYNRNDVAFTRGTFRVRGDTVEIIPSY
TH_1003|M.thermoresistible__bu RSVQISIGDEVPRDSLLRLLVDVQYTRNDTAFTRGSFRVRGDTVEIIPSY
MAB_2308|M.abscessus_ATCC_1997 RSVQLDVGLEVPRDALLRLLVDMQYNRNDMAFTRGTFRVRGDTVEIIPSY
***:: :. :****.*******:**.*** :****:**************
Mb1659|M.bovis_AF2122/97 EELAVRIEFFGDEIEALYYLHPLTGEVIRQVDSLRIFPATHYVAGPERMA
Rv1633|M.tuberculosis_H37Rv EELAVRIEFFGDEIEALYYLHPLTGEVIRQVDSLRIFPATHYVAGPERMA
MLBr_01387|M.leprae_Br4923 EELAVRIEFFGDEIEALYYLHPLTGEVIRQVDSLRIFPATHYVAGPERMA
MAV_3141|M.avium_104 EELAVRIEFFGDEIEALYYLHPLTGDVIRQVDSLRIFPATHYVAGPERMA
MMAR_2437|M.marinum_M EELAVRIEFFGDEIEALYYLHPLTGDVVRQVDSLRIFPATHYVAGPERMA
MUL_1616|M.ulcerans_Agy99 EDLAVRIEFFGDGIEALYYLHPLTGDVVRQVDSLRIFPATHYVAGPERMA
Mflv_3556|M.gilvum_PYR-GCK EELAVRIEFFGDEIEELYYLHPLTGDIIRKVDSLRIFPATHYVAGPERMA
Mvan_3350|M.vanbaalenii_PYR-1 EELAVRIEFFGDEIEELYYLHPLTGDIIRKVDSLRIFPATHYVAGPERMA
MSMEG_3816|M.smegmatis_MC2_155 EELAVRIEFFGDEIEALYYLHPLTGDVVRQVESLRIFPATHYVAGPERMA
TH_1003|M.thermoresistible__bu EELAVRIEFFGDEVEELYYLHPLTGDVVRKVDSVRIFPATHYVAGPERMA
MAB_2308|M.abscessus_ATCC_1997 EELAVRIEFFGDEVEALYYLHPLTGDVVRQTNSLRIFPATHYVAGPERMG
*:********** :* *********:::*:.:*:***************.
Mb1659|M.bovis_AF2122/97 HAVSAIEEELAERLAELESQGKLLEAQRLRMRTNYDIEMMRQVGFCSGIE
Rv1633|M.tuberculosis_H37Rv HAVSAIEEELAERLAELESQGKLLEAQRLRMRTNYDIEMMRQVGFCSGIE
MLBr_01387|M.leprae_Br4923 QAISAIEEELAERLAEFERQGKLLEAQRLRMRTNYDIEMMRQVGFCSGIE
MAV_3141|M.avium_104 HAISTIEQELAERLAELEGQGKLLEAQRLRMRTNYDIEMMRQVGFCSGIE
MMAR_2437|M.marinum_M QAISTIEEELAERLAELEGHGKLLEAQRLRMRTNYDIEMMRQVGFCSGIE
MUL_1616|M.ulcerans_Agy99 QAISTIEEELAERLAELEGHGKLLEAHRLRMRTNYDIEMMRQVGFCSGIE
Mflv_3556|M.gilvum_PYR-GCK AAISTIEAELEARLAELEGQGKLLEAQRLRMRTNYDIEMMRQVGFCSGIE
Mvan_3350|M.vanbaalenii_PYR-1 QAISTIEAELEERLAELEGQGKLLEAQRLRMRTNYDIEMMRQVGFCSGIE
MSMEG_3816|M.smegmatis_MC2_155 AAIESIEKELEERLAELENQGKLLEAQRLRMRTNYDVEMMRQVGFCSGIE
TH_1003|M.thermoresistible__bu QAISSIEDELAERLDELESQGKLLEAQRLRMRTNYDIEMMRQVGFCSGIE
MAB_2308|M.abscessus_ATCC_1997 KALEGIEQELEERLSELEGQGKLLEAQRLRMRTNYDIEMMRNVGFCSGIE
*:. ** ** ** *:* :******:*********:****:********
Mb1659|M.bovis_AF2122/97 NYSRHIDGRGPGTPPATLLDYFPEDFLLVIDESHVTVPQIGGMYEGDISR
Rv1633|M.tuberculosis_H37Rv NYSRHIDGRGPGTPPATLLDYFPEDFLLVIDESHVTVPQIGGMYEGDISR
MLBr_01387|M.leprae_Br4923 NYSRHIDGRGPGTPPATLLDYFPEDFLLVIDESHVTVPQIGGMYEGDMSR
MAV_3141|M.avium_104 NYSRHIDGRGPGSPPATLLDYFPEDFLLVIDESHVTVPQIGGMYEGDMSR
MMAR_2437|M.marinum_M NYSRHIDGRPAGSAPATLLDYFPEDFLLVIDESHVTVPQIGGMYEGDISR
MUL_1616|M.ulcerans_Agy99 NYSRHIDGRPAGSAPATLLDYFPEDFLLVIDESHVTVPQIGGMYEGDISR
Mflv_3556|M.gilvum_PYR-GCK NYSRHIDGRPAGSAPATLLDYFPEDFLLVIDESHVTVPQIGGMYEGDMSR
Mvan_3350|M.vanbaalenii_PYR-1 NYSRHIDGRPAGSAPATLLDYFPEDFLLVIDESHVTVPQIGGMYEGDMSR
MSMEG_3816|M.smegmatis_MC2_155 NYSRHIDGRPAGSAPATLLDYFPEDFLLVIDESHVTVPQIGGMYEGDMSR
TH_1003|M.thermoresistible__bu NYSRHIDGRPAGSAPATLLDYFPEDFLLVIDESHVTVPQIGGMYEGDISR
MAB_2308|M.abscessus_ATCC_1997 NYSRHIDGRGAGTPPATLLDYFPDDFLLVIDESHVTVPQIGAMYEGDMSR
********* .*:.*********:*****************.*****:**
Mb1659|M.bovis_AF2122/97 KRNLVEYGFRLPSACDNRPLTWEEFADRIGQTVYLSATPGPYELSQTGGE
Rv1633|M.tuberculosis_H37Rv KRNLVEYGFRLPSACDNRPLTWEEFADRIGQTVYLSATPGPYELSQTGGE
MLBr_01387|M.leprae_Br4923 KRNLVEYGFRLPSACDNRPLTWEEFADRIGQTVYLSATPGPYELSQSGGE
MAV_3141|M.avium_104 KRNLVEYGFRLPSACDNRPLTWEEFADRIGQTVYLSATPGPYELSQTGGE
MMAR_2437|M.marinum_M KRNLVEFGFRLPSAVDNRPLQWEEFADRIGQTVYLSATPGPYELSQSGGE
MUL_1616|M.ulcerans_Agy99 KRNLVEFGFRLPSAVDNRPLQWEEFADRIGQTVYLSATPGPYELSQSGGE
Mflv_3556|M.gilvum_PYR-GCK KRNLVDFGFRLPSAVDNRPLTWEEFADRIGQTVYLSATPGNFELAQTGGE
Mvan_3350|M.vanbaalenii_PYR-1 KRNLVDFGFRLPSAVDNRPLTWEEFADRIGQTVYLSATPGSYELSQSGGE
MSMEG_3816|M.smegmatis_MC2_155 KRNLVDFGFRLPSAVDNRPLTWEEFAERIGQTVYLSATPGPYEISQAGGE
TH_1003|M.thermoresistible__bu KRNLVEYGFRLPSAVDNRPLTWEEFAARIGQTVYLSATPGPYELSQSGGE
MAB_2308|M.abscessus_ATCC_1997 KRNLVDFGFRLPSAVDNRPLTWEEFADRIGQTVYLSATPGNYERSAAGGE
*****::******* ***** ***** ************* :* : :***
Mb1659|M.bovis_AF2122/97 FVEQVIRPTGLVDPKVVVKPTKGQIDDLIGEIRTRADADQRVLVTTLTKK
Rv1633|M.tuberculosis_H37Rv FVEQVIRPTGLVDPKVVVKPTKGQIDDLIGEIRTRADADQRVLVTTLTKK
MLBr_01387|M.leprae_Br4923 FVEQVIRPTGLVDPKVVVKPTKGQIDDLIGEIRKRANADQRVLVTTLTKK
MAV_3141|M.avium_104 FVEQVIRPTGLVDPKVVVKPTKGQIDDLIGEIRKRTEADERVLVTTLTKK
MMAR_2437|M.marinum_M FVEQVIRPTGLVDPKVVVKPTKGQIDDLIGEIRQRAEADQRVLVTTLTKK
MUL_1616|M.ulcerans_Agy99 FVEQVIRPTGLVDPKVVVKPTKGQIDDLIGEIRQRAEVDQRVLVTTLTKK
Mflv_3556|M.gilvum_PYR-GCK FVEQVIRPTGLVDPQVIVKPTKGQIDDLIGEIRKRTERDERVLVTTLTKK
Mvan_3350|M.vanbaalenii_PYR-1 FVEQVIRPTGLVDPQVVVKPTKGQIDDLIGEIRKRTERDERVLVTTLTKK
MSMEG_3816|M.smegmatis_MC2_155 FVEQVIRPTGLVDPKVVVKPTKGQIDDLIGEIRTRTERDERVLVTTLTKK
TH_1003|M.thermoresistible__bu VVEQVIRPTGLVDPKVVVKPTKGQIDDLIGEIRKRAERDERVLVTTLTKK
MAB_2308|M.abscessus_ATCC_1997 FVEQVIRPTGLVDPKVVVKPTKGQIDDLIASIRQRTEADERVLVTTLTKK
.*************:*:************..** *:: *:**********
Mb1659|M.bovis_AF2122/97 MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGDYDVLVGINLLR
Rv1633|M.tuberculosis_H37Rv MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGDYDVLVGINLLR
MLBr_01387|M.leprae_Br4923 MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGDYDVLVGINLLR
MAV_3141|M.avium_104 MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGEYDVLVGINLLR
MMAR_2437|M.marinum_M MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGEYDVLVGINLLR
MUL_1616|M.ulcerans_Agy99 MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGEYDVLVGINLLR
Mflv_3556|M.gilvum_PYR-GCK MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGEYDVLIGINLLR
Mvan_3350|M.vanbaalenii_PYR-1 MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGEYDVLVGINLLR
MSMEG_3816|M.smegmatis_MC2_155 MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGEYDVLVGINLLR
TH_1003|M.thermoresistible__bu MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGEYDVLVGINLLR
MAB_2308|M.abscessus_ATCC_1997 MAEDLTDYLLEMGIRVRYLHSEVDTLRRVELLRQLRLGEYDVLVGINLLR
**************************************:****:******
Mb1659|M.bovis_AF2122/97 EGLDLPEVSLVAILDADKEGFLRSSRSLIQTIGRAARNVSGEVHMYADKI
Rv1633|M.tuberculosis_H37Rv EGLDLPEVSLVAILDADKEGFLRSSRSLIQTIGRAARNVSGEVHMYADKI
MLBr_01387|M.leprae_Br4923 EGLDLPEVSLVAILDADKEGFLRSARSLIQTIGRAARNVSGEVHMYADTI
MAV_3141|M.avium_104 EGLDLPEVSLVAILDADKEGFLRSARSLIQTIGRAARNVSGEVHMYADTI
MMAR_2437|M.marinum_M EGLDLPEVSLVAILDADKEGFLRSARSLIQTIGRAARNVSGEVHMYADKI
MUL_1616|M.ulcerans_Agy99 EGLDLPEVSLVAILDADKEGFLRSARSLIQTIGRAARNVSGEVHMYADKI
Mflv_3556|M.gilvum_PYR-GCK EGLDLPEVSLVAILDADKEGFLRSPRSLIQTIGRAARNVSGEVHMYADKI
Mvan_3350|M.vanbaalenii_PYR-1 EGLDLPEVSLVAILDADKEGFLRSPRSLIQTIGRAARNVSGEVHMYADKM
MSMEG_3816|M.smegmatis_MC2_155 EGLDLPEVSLVSILDADKEGFLRSTRSLIQTIGRAARNVSGEVHMYADKI
TH_1003|M.thermoresistible__bu EGLDLPEVSLVSILDADKEGFLRSTRSLIQTIGRAARNVSGEVHMYADNI
MAB_2308|M.abscessus_ATCC_1997 EGLDLPEVSLVAILDADKEGFLRSSRSLIQTIGRAARNVSGEVHMYADTM
***********:************.***********************.:
Mb1659|M.bovis_AF2122/97 TDSMREAIDETERRRAKQIAYNEANGIDPQPLRKKIADILDQVYREADDT
Rv1633|M.tuberculosis_H37Rv TDSMREAIDETERRRAKQIAYNEANGIDPQPLRKKIADILDQVYREADDT
MLBr_01387|M.leprae_Br4923 TDSMTEAIDETERRRAKQIAYNNANGIDPQPLRKKIADILDQVYREADDT
MAV_3141|M.avium_104 TESMKEAIDETERRRAKQIAYNEAHGIDPQPLRKKIADILDQVYREADDT
MMAR_2437|M.marinum_M TDSMKEAIDETERRRAKQVAYNEANGIDPQPLRKKIADILDQVYREADDT
MUL_1616|M.ulcerans_Agy99 TDSMKEAIDETERRRAKQVAYNEANGIDPQPLRKKIADILDQVYREADDT
Mflv_3556|M.gilvum_PYR-GCK TDSMKQAIDETERRRAKQIAYNTERGIDPQPLRKKIADILDQVYREADDT
Mvan_3350|M.vanbaalenii_PYR-1 TDSMKQAIDETERRRAKQTAYNKEHGIDPKPLRKKIADILDQVYREADDT
MSMEG_3816|M.smegmatis_MC2_155 TDSMREAIDETERRRAKQIAYNEANGIDPKPLRKKIADILDQVYREADDT
TH_1003|M.thermoresistible__bu TDSMREAIDETERRRAKQIAYNEKHGIDPQPLRKKIADILDQVYREAEDT
MAB_2308|M.abscessus_ATCC_1997 TDSMKEAIDETDRRRAKQIAYNQANGIDPQPLRKKIADILDQVYREAEDT
*:** :*****:****** *** .****:*****************:**
Mb1659|M.bovis_AF2122/97 ---AVVEVGGSGRNASRGRRAQGEPGRAVSAGVFEGRDTSAMPRAELADL
Rv1633|M.tuberculosis_H37Rv ---AVVEVGGSGRNASRGRRAQGEPGRAVSAGVFEGRDTSAMPRAELADL
MLBr_01387|M.leprae_Br4923 ---DTVQVGGSGRNVSRGRRAQSEPVRSVSVGVFEGRDTAGMPRAELADL
MAV_3141|M.avium_104 ---ETVEIG-SGRSMSRGRRAQGEPGRAVSAGIVEGRDTTNMPRAELADL
MMAR_2437|M.marinum_M EAAESVPIGGSGRNSSRGRRAQGEPGRAVSAGVFEGRDTSAMPRAELADL
MUL_1616|M.ulcerans_Agy99 EAAESVPIGGSGRNSSRGRRAQGEPGRAVSAGVFEGRDTSTMPRAELADL
Mflv_3556|M.gilvum_PYR-GCK EAVE---IGGSGRNASRGRRAPGEPGRAVSAGVYEGRDTKNMPRAELADL
Mvan_3350|M.vanbaalenii_PYR-1 EAAESVPIGGSGRNASRGRRAQGEPGRAVSAGVFEGRDTSNMPRAELADL
MSMEG_3816|M.smegmatis_MC2_155 ESVE---IGGSGRNSSRGRRAQGEPGRAVSAGIVEGKDTSNMPRAELADL
TH_1003|M.thermoresistible__bu ETVE---IG-SGRAVSRGRRAQGEPGRGVTAGIIEGRDTRNMPRAELADL
MAB_2308|M.abscessus_ATCC_1997 ---EEVAVGGSGRNSSRGRRAQGEPGRAVSAGVFEGRDTKTMPRAELADL
:* *** ****** .** *.*:.*: **:** *********
Mb1659|M.bovis_AF2122/97 IKDLTAQMMAAARDLQFELAARFRDEIADLKRELRGMDAAGLK
Rv1633|M.tuberculosis_H37Rv IKDLTAQMMAAARDLQFELAARFRDEIADLKRELRGMDAAGLK
MLBr_01387|M.leprae_Br4923 IKDLTAQMMAAASDLQFELAARFRDEIADLKKELRGMDAAGLK
MAV_3141|M.avium_104 IKDLTAQMMAAARDLQFELAARFRDEIADLKKELRGMDAAGLK
MMAR_2437|M.marinum_M IKDLTSQMMVAARDLQFELAARFRDEIADLKKELRGMDAAGLK
MUL_1616|M.ulcerans_Agy99 IKDLTSQMMVAARDLQFELAARFRDEIADLKKELRGMDAAGLN
Mflv_3556|M.gilvum_PYR-GCK IKDLTAQMMAAARDLQFELAARIRDEIQDLKKELRGMDAAGLK
Mvan_3350|M.vanbaalenii_PYR-1 IKDLTAQMMAAARDLQFELAARIRDEIADLKKELRGMDAAGLK
MSMEG_3816|M.smegmatis_MC2_155 IKELTDQMMAAARDLQFELAARIRDEIADLKKELRGMDAAGLK
TH_1003|M.thermoresistible__bu IKELTEQMMAAARDLQFEVAARVRDEIADLKKELRGMDAAGLK
MAB_2308|M.abscessus_ATCC_1997 IKDMTEQMMTAARDLQFELAARFRDEIADLKKELRGMDAAGLK
**::* ***.** *****:***.**** ***:**********: