For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQPVTERDAGRDCPKRMTFGPCGGVHPDGQCEMRPGPCAFPDIVPWPTQAAPPPVTAPLILTDFSCTPFD PHDVAATANALAPSCDAVLVGEHQNRPDFPPTLMSRLLLEAGVTPWITLSCRDRNRVVLEQELRGLSLLG VQTVLCVTGDGRAYDVRPDVTQTFDLDGPRLVELAASLGMVAAVPETPTAPPVARRPLRLVQKQHAGAGI AVLNHVSSPSTVAEFMAAARAAGLVIPVLAAVAVFTDDVSAAVLQGLPGLALDPAVVDRVLAAPDPVEAG IEAAVAEARALLAIDGVAGVNVSGLASGAGTRVGAQIKAEVGARIRAERSVR*
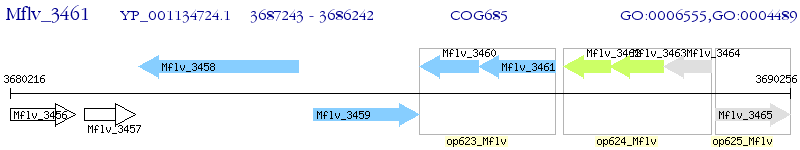
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3461 | - | - | 100% (333) | methylenetetrahydrofolate reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6664 | - | 1e-113 | 77.49% (271) | methylenetetrahydrofolate reductase family protein, putative |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3461|M.gilvum_PYR-GCK MTQPVTERDAGRDCPKRMTFGPCGGVHPDGQCEMRPGPCAFPDIVPWPTQ
MSMEG_6664|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3461|M.gilvum_PYR-GCK AAPPPVTAPLILTDFSCTPFDPHDVAATANALAPSCDAVLVGEHQNRPDF
MSMEG_6664|M.smegmatis_MC2_155 ----------MLTDFSCAPFDEQDVVRTAGVLAGSCDAVLVGEHQNKPDF
:******:*** :**. **..** ************:***
Mflv_3461|M.gilvum_PYR-GCK PPTLMSRLLLEA----GVTPWITLSCRDRNRVVLEQELRGLSLLGVQTVL
MSMEG_6664|M.smegmatis_MC2_155 PPTLMGRLLLDALTGSGVTPWITLSCRDRNRVVLEQELRGLRSIGADTVM
*****.****:* ************************* :*.:**:
Mflv_3461|M.gilvum_PYR-GCK CVTGDGRAYDVRPDVTQTFDLDGPRLVELAASLGMVAAVPETPTAPPVAR
MSMEG_6664|M.smegmatis_MC2_155 CVTGDGRGYDVRPDVTQTFDLDGPRLVALAASLDMVAAVPETPTAPPVNA
*******.******************* *****.**************
Mflv_3461|M.gilvum_PYR-GCK RPLRLVQKQHAGAGIAVLNHVSSPSTVAEFMAAARAAGLVIPVLAAVAVF
MSMEG_6664|M.smegmatis_MC2_155 RPRRLVEKQKAGAGIAVLNHVPYPHTVARFMESGRAAGLTIPVIAAVAVF
** ***:**:***********. * ***.** :.*****.***:******
Mflv_3461|M.gilvum_PYR-GCK TDDVSAAVLQGLPGLALDPAVVDRVLAAPDPVEAGIEAAVAEARALLAID
MSMEG_6664|M.smegmatis_MC2_155 TDTASAAVLQGLPGLELDPAVTQRVLGAADPVEAGIEAAVEEARALLSID
** .*********** *****.:***.*.*********** ******:**
Mflv_3461|M.gilvum_PYR-GCK GVAGVNVSGLASGAGTRVGAQIKAEVGARIR---AERSVR---
MSMEG_6664|M.smegmatis_MC2_155 GIEGVNISGSASASGG--GAAIKAEVGRRVKNLCVDRTRGEAP
*: ***:** **.:* ** ****** *:: .:*: