For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
METFTPTLDWGNELWTSLWWIAQGWVYAAIATMVVLTLIVRFTTWGRQFWRVTRGYFTGRDSVVVWLWLA GILLLVIASVRLSVLFSFQGNDMMTSFQVIASGVGAGDDAVRASGRDGFWLSMLVFSVLAVVNVVTIMID LLVTQRFMLRWRAWLTDQLTGDWLDGRAYYRSRFIDDSIDNPDQRIQMDIDIFTTGVGPLPNRPNNTSGT TLLFGAISSIAAMISFTTILWNLSGPVTLPFIGYELPKAMFWIGIVYILFATVVAFWIGRPIITLAFRNE KFNAAFRYALVRLRDAAEAVAFYRGEIAERTGLRKLFAPVVDNYKRYVNRMAGFLGWNLSITQAQELIPY IVQFSRFYNGEITLGQLSQTASAFREILSGLSFFRNAYDDFAGYRAAIIRLHGLVVANEEGRALPSLDTQ DSTDGRVELDDVEVRTPDGRQLLQPVDLRLEPGDGLVITGPSGSGKTTLLRSLGRLWPFASGTLVYPAGE NDTMFLSQLPYVPLGDLRAVVSYPNEPGSIPDGTLREVLEKVALPHLADRLDEEQDWAKVLSPGEQQRVA FARILLTKPEAVFLDESTSALDEGLELMLYRLVRSELPQTIVVSVSHRSTVEQHHSRQLKLLGDGRWEVG AVQSR
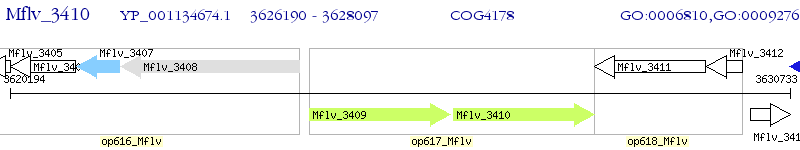
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3410 | - | - | 100% (635) | ABC transporter |
| M. gilvum PYR-GCK | Mflv_3409 | - | 0.0 | 53.50% (628) | ABC transporter |
| M. gilvum PYR-GCK | Mflv_2250 | - | 2e-19 | 29.32% (266) | ABC transporter, transmembrane region |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1850c | - | 0.0 | 61.33% (631) | drugs-transport transmembrane ATP-binding protein ABC |
| M. tuberculosis H37Rv | Rv1819c | - | 0.0 | 61.17% (631) | drugs-transport transmembrane ATP-binding protein ABC |
| M. leprae Br4923 | MLBr_02084 | - | 0.0 | 60.51% (633) | putative multidrug resistance pump |
| M. abscessus ATCC 19977 | MAB_3399 | - | 1e-180 | 51.21% (621) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_2696 | - | 0.0 | 62.00% (629) | drug-transport transmembrane ATP-binding protein ABC |
| M. avium 104 | MAV_2896 | - | 0.0 | 60.48% (630) | SbmA protein |
| M. smegmatis MC2 155 | MSMEG_3655 | - | 0.0 | 75.40% (630) | ABC transporter, permease/ATP-binding protein |
| M. thermoresistible (build 8) | TH_2327 | - | 0.0 | 63.00% (627) | PROBABLE DRUGS-TRANSPORT TRANSMEMBRANE ATP-BINDING PROTEIN ABC |
| M. ulcerans Agy99 | MUL_3059 | - | 0.0 | 61.37% (629) | drug-transport transmembrane ATP-binding protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3164 | - | 0.0 | 81.75% (630) | ABC transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3410|M.gilvum_PYR-GCK --METFTPTLDWGNELWTSLWWIAQGWVYAAIATMVVLTLIVRFTTWGRQ
Mvan_3164|M.vanbaalenii_PYR-1 --METFTPTLDWGNELWTSLWWIAQGWAYAAVATIVVLALIVRFTTWGRQ
MSMEG_3655|M.smegmatis_MC2_155 --MGMFSPTLDWSNELLTSLIWIAKGWAIAAVCTLAVLALLVRFTTWGRQ
Mb1850c|M.bovis_AF2122/97 MGPKLFKPSIDWSRAFPDSVYWVGKAWTISAICVLAILVLLRYLTPWGRQ
Rv1819c|M.tuberculosis_H37Rv MGPKLFKPSIDWSRAFPDSVYWVGKAWTISAICVLAILVLLRYLTPWGRQ
MMAR_2696|M.marinum_M MGPKPFTPSIDWSSALVDSVFWVAKAWAISAVCVIAALALFKFFTPWGRQ
MUL_3059|M.ulcerans_Agy99 MGPKPFTPSIDWSSAFVDSVFWVAKAWAISAVCVIAALALFKFFTPWGRQ
MAV_2896|M.avium_104 MGAKPFKPSIDWSAAFVDSLRWLAIAWVISAVCLFGVLLAFRFLTPWGRQ
MLBr_02084|M.leprae_Br4923 MELRPFYPSVNWGYALPDSLWWIAKAWAISAACVV-VLVMLSFITRWGRQ
TH_2327|M.thermoresistible__bu VELEPFSPSIDWAGEWWESLKFIALTWTISAAGVLLICVLLGLFTVWGRQ
MAB_3399|M.abscessus_ATCC_1997 -----MKTATDWGTELWRSLVWIGWVWPLTAAIFVTIAFLIIQFTQWGRQ
: .: :*. *: ::. * :* . : :* ****
Mflv_3410|M.gilvum_PYR-GCK FWRVTRGYFTGRDSVVVWLWLAGILLLVIASVRLSVLFSFQGNDMMTSFQ
Mvan_3164|M.vanbaalenii_PYR-1 FWRVTRGYFTGPESLIVWLWLAVILLLVILSVRLSVLFSFQGNDMMTSFQ
MSMEG_3655|M.smegmatis_MC2_155 YWRITSGYFTGADSIKVWVWLAALLLSVITGVRLSVLFTYQGNDMMTSFQ
Mb1850c|M.bovis_AF2122/97 FWRITRAYFVGPNSVRVWLMLGVLLLSVVLAVRLNVLFSYQGNDMYTALQ
Rv1819c|M.tuberculosis_H37Rv FWRITRAYFVGPNSVRVWLMLGVLLLSVVLAVRLNVLFSYQGNDMYTALQ
MMAR_2696|M.marinum_M FWRITSEYFIGPKSIRVWLMLGVLLLSVVLAVRLNVLFSYQGNDMYTALQ
MUL_3059|M.ulcerans_Agy99 FWRITSEYFIGPKSIRVWLMLGVLLFSVVLAVRLNVLFSYQGNDMYTALQ
MAV_2896|M.avium_104 FWRITRGYFVGAHSVRVWLMLGVLLLSVLLSVRLSVLLSFQSNDLYTALQ
MLBr_02084|M.leprae_Br4923 YWRITRGYFIGPQSVGVWLMLAVLLFSVVISVRLNVLFSYQSKDLYNALQ
TH_2327|M.thermoresistible__bu FWRITGGYFVGRDSLPVWGLFAVLLLSVVASVRLNVLLSYYSNDLYSALQ
MAB_3399|M.abscessus_ATCC_1997 FWRITGQYFTTRAGRGGLVLLGLVLLSVVVNVRLSVVFTYYYNDMSASIQ
:**:* ** . :. :*: *: ***.*:::: :*: ::*
Mflv_3410|M.gilvum_PYR-GCK VIASGVGAGDDAVRASGRDGFWLSMLVFSVLAVVNVVTIMIDLLVTQRFM
Mvan_3164|M.vanbaalenii_PYR-1 VIASGVGAGDDAVKQSGGDGFWMSMGVFAVLAVINVVSIMVDLLLTQRFM
MSMEG_3655|M.smegmatis_MC2_155 VIAEGVASGDEAVRTSGKDGFWMSLTVFTILAALNVGRIMLDLFMAQRFM
Mb1850c|M.bovis_AF2122/97 KAFEGIASGDGTVKRSGARGFWMSIGVFSVMAVLHVTRVMADIYLTQRFI
Rv1819c|M.tuberculosis_H37Rv KAFEGIASGDGTVKRSGVRGFWMSIGVFSVMAVLHVTRVMADIYLTQRFI
MMAR_2696|M.marinum_M KAFQGTAASDEAVKRSGVRGFWMSIGVFSVMAVIHVARVMADIYLTQRFI
MUL_3059|M.ulcerans_Agy99 KAFQGTAASDEAVKRSGVRGFWMSIGVFSVMAVTHVARVMADIYLTQRFI
MAV_2896|M.avium_104 KAFEGIASGNDAVKHSGIHGFWVSLGIFCLLAALWVTRFMADVYLTQRFI
MLBr_02084|M.leprae_Br4923 AAFEGAGAQNDLVKQSGMHGFWMSLGIFSILAVIFIARVMADIYLTQRFI
TH_2327|M.thermoresistible__bu SAFQGAASGDEAVRDSGVSGFWFAIWVFVVLATVHVARTMLDIYLMQRFI
MAB_3399|M.abscessus_ATCC_1997 YIVQALARDGQGMP-EAKAFFWLNIRRFCLLAGINIAVVVTDFYLLQAFI
.. . . : .. **. : * ::* : : *. : * *:
Mflv_3410|M.gilvum_PYR-GCK LRWRAWLTDQLTGDWLDGRAYYRSRFIDDSIDNPDQRIQMDIDIFTTGVG
Mvan_3164|M.vanbaalenii_PYR-1 LRWRTWLTDRLTGDWLDGKAYYRSRFIDDTIDNPDQRIQADIDIFTAGVG
MSMEG_3655|M.smegmatis_MC2_155 LAWRQWLTARLTSDWLDGKAYYRSRFIDDTIDNPDQRIQTDIDIFTAGVG
Mb1850c|M.bovis_AF2122/97 IAWRVWLTHHLTQDWLDGRAYYRDLFIDETIDNPDQRIQQDVDIFTAGAG
Rv1819c|M.tuberculosis_H37Rv IAWRVWLTHHLTQDWLDGRAYYRDLFIDETIDNPDQRIQQDVDIFTAGAG
MMAR_2696|M.marinum_M IAWRVWLTDHLTKDWLKGRAYYRDLFIDETIDNPDQRIQQDIDVFTAGAG
MUL_3059|M.ulcerans_Agy99 IAWRVWLTDHLTKDWLKGRAYYRDLFIDETIDNPDQRIQQDIDVFTAGAG
MAV_2896|M.avium_104 IAWRMWLTANLTDDWLAGRAYYRDLFIDNTIDNPDQRIQQDIDIFTANAG
MLBr_02084|M.leprae_Br4923 IAWRMWLTNHLTTDWLDQRAYYREQFIDNTIDNPDQRIQQDIDIFTAGAG
TH_2327|M.thermoresistible__bu IRWRVWLTDRLTTDWLDNHAYYRARFIDRTIDNPDQRIQTDIDIFTTGYG
MAB_3399|M.abscessus_ATCC_1997 IRWRVWLTERITVDWLSKRAFYRSRFIDNTIDNPDQRIQADINNFVAMSS
: ** *** .:* *** :*:** *** :********* *:: *.: .
Mflv_3410|M.gilvum_PYR-GCK PLPNRPNNTSGTTLLFGAISSIAAMISFTTILWNLSGPVTLPFIGYELPK
Mvan_3164|M.vanbaalenii_PYR-1 PLPNMPNNTSGSTLLFGAVSSIAAMISFTTILWNLSGPVTLPFVGFELPK
MSMEG_3655|M.smegmatis_MC2_155 PTPNTPNNTSTATLLFGAVNAIASMISFTAILWNLSGNLTLPLIGVELPR
Mb1850c|M.bovis_AF2122/97 GTPNAPSNGTASTLLFGAVQSIISVISFTAILWNLSG--TLNIFGVSIPR
Rv1819c|M.tuberculosis_H37Rv GTPNAPSNGTASTLLFGAVQSIISVISFTAILWNLSG--TLNIFGVSIPR
MMAR_2696|M.marinum_M GTPNQPSNGTASTLLFGAVQSIISVISFTAILWNLSG--TLNIFGLSFPR
MUL_3059|M.ulcerans_Agy99 GTPNQPSNGTASMLLFGAVQSIISVISFTAILWNLSG--TLNIFGLSFPR
MAV_2896|M.avium_104 GTPNVPSNGTGSTLLFGAVNAVASVISFTAILWKLSG--DLNLFGLVLPR
MLBr_02084|M.leprae_Br4923 GTPNGPSNGTGSTLLFGAVESVISVISFTAILWNLSG--RLAVFEFEIPR
TH_2327|M.thermoresistible__bu TTPNVPSYGTGTVLLFGAVESVVSVVSFAAILWNLSGPLSVPLIDLTIPR
MAB_3399|M.abscessus_ATCC_1997 APEQLQS--SSLHLAFGAVNACISLPSFTIILWNLSG--PLNIFGYEMPR
: . : * ***:.: :: **: ***:*** : .. :*:
Mflv_3410|M.gilvum_PYR-GCK AMFWIGIVYILFATVVAFWIGRPIITLAFRNEKFNAAFRYALVRLRDAAE
Mvan_3164|M.vanbaalenii_PYR-1 AMFVIGIVYILFATVVAFVIGRPIITLSFNNEKFNAAFRYALVRLRDAAE
MSMEG_3655|M.smegmatis_MC2_155 AMFFIGILYVVVATVIAFWIGRPIIWLAFDNEKFNAAFRYALVRMRDAAE
Mb1850c|M.bovis_AF2122/97 AMFWTVLVYVFVATVISFIIGRPLIWLSFRNEKLNAAFRYALVRLRDAAE
Rv1819c|M.tuberculosis_H37Rv AMFWTVLVYVFVATVISFIIGRPLIWLSFRNEKLNAAFRYALVRLRDAAE
MMAR_2696|M.marinum_M AMFWTVLIYVLVATIISFVIGRPLIWLSFRNEKLNAAFRYALVRLRDAAE
MUL_3059|M.ulcerans_Agy99 AMFWTVLIYVLVATIISFVIGRPLIWLSFRNEKLNAAFRYALVRLRDAAE
MAV_2896|M.avium_104 AMFWTVLVYVFIATVVAVWLGRPLIWLSFNNEKLNAAFRYALVRLRDAAE
MLBr_02084|M.leprae_Br4923 AMFWIVIVYVLLATIITFWIGRPLIGLNFANEKLNAAFRYALVRLRDAAE
TH_2327|M.thermoresistible__bu ALFWIAIAFVIFATVVAFWIGRPLIRLSFRNEAFNAAFRYALVRLRDAAE
MAB_3399|M.abscessus_ATCC_1997 ALVFLTYIYVIVTTLIAFRIGRPLIRRFFLNERLNAMFRYALVRLRDTSE
*:. ::..:*:::. :***:* * ** :** *******:**::*
Mflv_3410|M.gilvum_PYR-GCK AVAFYRGEIAERTGLRKLFAPVVDNYKRYVNRMAGFLGWNLSITQAQELI
Mvan_3164|M.vanbaalenii_PYR-1 AVAFYRGEIAERTGLRRRFGPIVENYKKYVNRMAGFLGWNLSISQAQELI
MSMEG_3655|M.smegmatis_MC2_155 AVAFYRGEFAERTGLRRLFAPVVSNYKRYINRMAKFYGWNLSVSQAQELI
Mb1850c|M.bovis_AF2122/97 AVGFYRGERVEGTQLQRRFTPVIDNYRRYVRRSIAFNGWNLSVSQTIVPL
Rv1819c|M.tuberculosis_H37Rv AVGFYRGERVEGTQLQRRFTPVIDNYRRYVRRSIAFNGWNLSVSQTIVPL
MMAR_2696|M.marinum_M AVGFYRGERVEGAQLERRFLPVIRNYRRYVRRSIAFNGWNLSVSQTIVPL
MUL_3059|M.ulcerans_Agy99 AVGFYRGERVEGAQLERRFLPVIRNYRRYVRRSIAFNGWNLSVSQTIVPL
MAV_2896|M.avium_104 AVGFYRGERVERAQLWQRFTPIIANYRRYVRRTIIFNGWNWSVTQIIVPL
MLBr_02084|M.leprae_Br4923 AVAFYRGEEAERKQLEQRFTPIIDNYRRYVSRTIRFLGWNASVSQTIVPL
TH_2327|M.thermoresistible__bu AVGFYRGERAEKVELDRRFNATILNYRRWVARTIGFTGWNLTMSQAINPL
MAB_3399|M.abscessus_ATCC_1997 SVAFYRGERAESKQLKSRFDAIIANFWQLVYRSMGFTGWNFAVSQTAAVF
:*.***** .* * * . : *: : : * * *** :::* :
Mflv_3410|M.gilvum_PYR-GCK PYIVQFSRFYNGEITLGQLSQTASAFREILSGLSFFRNAYDDFAGYRAAI
Mvan_3164|M.vanbaalenii_PYR-1 PYIVQFPRFFSGEITLGQLSQTAGAFREILTGLSFFRNAYDQFAGYRAAI
MSMEG_3655|M.smegmatis_MC2_155 PYIVQFPRFFNGEVTLGGLTQTASAFRNLMDGLSFFRNAYDQFAGYRAAI
Mb1850c|M.bovis_AF2122/97 PWVIQAPRLFAGQIDFGDVGQTATSFGNIHDSLSFFRNNYDAFASFRAAI
Rv1819c|M.tuberculosis_H37Rv PWVIQAPRLFAGQIDFGDVGQTATSFGNIHDSLSFFRNNYDAFASFRAAI
MMAR_2696|M.marinum_M PWVIQAPRLFAGQIDFGDVGQTATAFGNIHDSLSFFRNNYDAFASFRAAI
MUL_3059|M.ulcerans_Agy99 PWVIQAPRLFAGQIDFGDVGQTATAFGNIHDSLSFFRNNYDAFASFRAAI
MAV_2896|M.avium_104 PLAIQAPRLFAGEIAFGDVTQTATAFNNIHDSLSFFRNNYDAFAAFRAAI
MLBr_02084|M.leprae_Br4923 PWILQAPRLFAGKINLGDVGQTATAFGNIHDSLSFFRNNYAAFAAFRAAI
TH_2327|M.thermoresistible__bu PIVVQAPRLFNGEIALGAVMQSASAFGEIQNGLSFFRNAYDNFASYRAAI
MAB_3399|M.abscessus_ATCC_1997 NSVLQVPRLIAGQITYGDVSQSASAVGQLQTSLSFFRNAYDNFAAYRATV
:* .*: *:: * : *:* :. :: .****** * **.:**::
Mflv_3410|M.gilvum_PYR-GCK IRLHGLVVANEEGRALPSLDTQDSTDGRVELDDVEVRTPDGRQLLQPVDL
Mvan_3164|M.vanbaalenii_PYR-1 IRLHGLVVANEEGRELPTLTVEPSSGNTVELDDVEVRKPDGTQLIDPIDL
MSMEG_3655|M.smegmatis_MC2_155 IRLHGLVVANEEARALPEVTTTACVDGTVELKDVEVRTPDGRQLIKPLDM
Mb1850c|M.bovis_AF2122/97 IRLHGLVDANEKGRALPAVLTRPSDDESVELNDIEVRTPAGDRLIDPLDV
Rv1819c|M.tuberculosis_H37Rv IRLHGLVDANEKGRALPAVLTRPSDDESVELNDIEVRTPAGDRLIDPLDV
MMAR_2696|M.marinum_M IRLHGLVDANEKGRALPAVLTKPSSDESVELTDVEVRTPAGDRLIDPLDV
MUL_3059|M.ulcerans_Agy99 IRLHGLVDANEKGRALPAVLTKPSSDESVELTDVEVRTPAGDRLIDPLDV
MAV_2896|M.avium_104 IRLHGLVDANSKGRALPAILVKPSEETAVELRGIEVRTPEGDQLVDSLDI
MLBr_02084|M.leprae_Br4923 IRLHGLVVANARSRELPHVTAKSTNDGTVHVVAVEVRTSAGEQLIDPLDL
TH_2327|M.thermoresistible__bu IRLHGLVEANEAARAMGALETAASTDGSVELSDVEVRTPTGRQLIEPLNL
MAB_3399|M.abscessus_ATCC_1997 MRLDGLLTADSQSRELPMLEPGDQEDG-VTLNNIYVRKPNGDSLIDDLNL
:**.**: *: .* : : * : : **.. * *:. :::
Mflv_3410|M.gilvum_PYR-GCK RLEPGDGLVITGPSGSGKTTLLRSLGRLWPFASGTLVYPAGENDTMFLSQ
Mvan_3164|M.vanbaalenii_PYR-1 RLEAGDTLAVTGVSGTGKTTLLRSLAQLWPYTTGTLRCPQGTNETMFLSQ
MSMEG_3655|M.smegmatis_MC2_155 RLEVGDTLVITGPSGSGKTTLLRSLAELWPFTSGTLTRPCGPNETMFLSQ
Mb1850c|M.bovis_AF2122/97 RLDRGGSLVITGRSGAGKTTLLRSLAELWPYASGTLHRPGGENETMFLSQ
Rv1819c|M.tuberculosis_H37Rv RLDRGGSLVITGRSGAGKTTLLRSLAELWPYASGTLHRPGGENETMFLSQ
MMAR_2696|M.marinum_M RLDRGDSLVITGRSGSGKTTLLRSLAELWPFASGTLSRPDGANDTMFLSQ
MUL_3059|M.ulcerans_Agy99 RLDRGDSLVITGRSGSGKTTLLRSLAELWPFASGTLSRPDGANDTMFLSQ
MAV_2896|M.avium_104 QLDEGDTLVITGRSGAGKTTLLRSLAELWPYASGTLCRPDGDNATMFLSQ
MLBr_02084|M.leprae_Br4923 QLNCGSSLVITGRSGVGKTTMLRSLAELWPYASGTLSRPDGKNETMFLSQ
TH_2327|M.thermoresistible__bu RLERGESLVITGRSGAGKTTLLRSLAQLWPYATGTLRRP-ADRDTMFLSQ
MAB_3399|M.abscessus_ATCC_1997 DLAAGDALLIKGQSGSGKTTLLRSLAGLWPYTDGDWARPGGDHETMFISQ
* * * :.* ** ****:****. ***:: * * . . ***:**
Mflv_3410|M.gilvum_PYR-GCK LPYVPLGDLRAVVSYPNEPGSIPDGTLREVLEKVALPHLADRLDEEQDWA
Mvan_3164|M.vanbaalenii_PYR-1 LPYVPLGDLRAVLSYPRQVGDIPDSELVAVLNKVALPHLVSRLDEEQDWA
MSMEG_3655|M.smegmatis_MC2_155 MPYVPLGDLRAVVTYPAPEGSVSDEKLAHTLNKVALPHLVERLDEVQDWA
Mb1850c|M.bovis_AF2122/97 LPYVPLGTLRDVVCYPNSAAAIPDATLRDTLTKVALAPLCDRLDEERDWA
Rv1819c|M.tuberculosis_H37Rv LPYVPLGTLRDVVCYPNSAAAIPDATLRDTLTKVALAPLCDRLDEERDWA
MMAR_2696|M.marinum_M LPYVPLGSLRVVVCYPNSPYDIPDEAVRDTLTKVALAPLCDRLDEERDWA
MUL_3059|M.ulcerans_Agy99 LPYVPLGSLRVVVCYPNSPYDIPDEAVRDTLTKVALAPLCDRLDEERDWA
MAV_2896|M.avium_104 LPYVPLGTLRTVVCYPNSPDSIADSQLHEVLTKVALAPLISRLDEDQDWA
MLBr_02084|M.leprae_Br4923 LPYVPLGNLRGVVCYPNAPADIPEDTVRDTLTKVALNPLCDRLDEERDWA
TH_2327|M.thermoresistible__bu LPYVPLGDLRTVVSYPAESGQIPDAELQRALHDVSLGHLVHRLDELQDWA
MAB_3399|M.abscessus_ATCC_1997 LPYLPLGNLRDALSYPAESGSFSDEELLQTLEKVSLAHLADRLDEEADWI
:**:*** ** .: ** ..: : .* .*:* * **** **
Mflv_3410|M.gilvum_PYR-GCK KVLSPGEQQRVAFARILLTKPEAVFLDESTSALDEGLELMLYRLVRSELP
Mvan_3164|M.vanbaalenii_PYR-1 KVLSPGEQQRVAFARILLTRPKAAFFDEATSALDVGLETMLYQMVRDELP
MSMEG_3655|M.smegmatis_MC2_155 KVLSPGEQQRIAFARILLTKPKVVFLDESTSALDEGLEFLLYQLVRTELP
Mb1850c|M.bovis_AF2122/97 KVLSPGEQQRVAFARILLTKPKAVFLDESTSALDTGLEFALYQLLRSELP
Rv1819c|M.tuberculosis_H37Rv KVLSPGEQQRVAFARILLTKPKAVFLDESTSALDTGLEFALYQLLRSELP
MMAR_2696|M.marinum_M KVLSPGEQQRVAFARILLTKPKAVFLDESTSALDTGLEFALYELLRNELP
MUL_3059|M.ulcerans_Agy99 KVLSPGEQQRAAFARILLTKPKAVFLDESTSALDTGLEFALYELLRNELP
MAV_2896|M.avium_104 KVLSPGEQQRVAFARVLLTRPKAVFLDEATSALDEGLEYALYQLVRAELP
MLBr_02084|M.leprae_Br4923 KILSPGEQQRIAFARILLTKPKAIFLDESTSALDEGLEFALYQMLRSELP
TH_2327|M.thermoresistible__bu KVLSPGEQQRVAFARVLLNRPQVVFLDEATSALDEGLEFQMYELLRDRLP
MAB_3399|M.abscessus_ATCC_1997 KVLSPGEQQRVAFARILLIKPKAVFMDESTSSLDEGLEFSLYRLLRSELP
*:******** ****:** :*:. *:**:**:** *** :*.::* .**
Mflv_3410|M.gilvum_PYR-GCK QTIVVSVSHRSTVEQHHSRQLKLLGDGRWEVG----AVQSR---------
Mvan_3164|M.vanbaalenii_PYR-1 DTILVSVAHRGTVIRQCEKELALLGDGRWRLG----LAGADAP-------
MSMEG_3655|M.smegmatis_MC2_155 DTILVSVSHRTTVEQHHTHELQLLGDGDWRLG----RTNENPKQALV---
Mb1850c|M.bovis_AF2122/97 DCIVVSVSHRPALERLHENQLELLGGGQWRLA----PVEAAPAEV-----
Rv1819c|M.tuberculosis_H37Rv DCIVISVSHRPALERLHENQLELLGGGQWRLA----PVEAAPAEV-----
MMAR_2696|M.marinum_M DCIVVSVSHRPALERLHNQQLELLGGGPWRLG----PVNQEPAPA-----
MUL_3059|M.ulcerans_Agy99 DCIVVSVSHRPALERLHNQQLELLGGGPWRLG----PVNQEPAPA-----
MAV_2896|M.avium_104 GCVMVSVSHRPTVEQHHDQQLHLLGGGPWQLS----PVEKEPARV-----
MLBr_02084|M.leprae_Br4923 DCIIVSVSHRRAVELLHAQQLELLGGGQWRLG----PVEKEPLHV-----
TH_2327|M.thermoresistible__bu DTILVSVSHRPTVEQHHHRHLELLGDGQWRXXXHTNRIGTSGQRVMCQYA
MAB_3399|M.abscessus_ATCC_1997 GCTLVSVGHRSTIDQHHNQKLQLDAEGRWSLS----PL------------
::**.** :: ..* * . * *
Mflv_3410|M.gilvum_PYR-GCK --------------------------------------
Mvan_3164|M.vanbaalenii_PYR-1 --------------------------------------
MSMEG_3655|M.smegmatis_MC2_155 --------------------------------------
Mb1850c|M.bovis_AF2122/97 --------------------------------------
Rv1819c|M.tuberculosis_H37Rv --------------------------------------
MMAR_2696|M.marinum_M --------------------------------------
MUL_3059|M.ulcerans_Agy99 --------------------------------------
MAV_2896|M.avium_104 --------------------------------------
MLBr_02084|M.leprae_Br4923 --------------------------------------
TH_2327|M.thermoresistible__bu LANTAQADSSGTSSPSTSTITARTSASAAVTMQSRARR
MAB_3399|M.abscessus_ATCC_1997 --------------------------------------