For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLTPHEQERLLLSYAAELARRRQARGLKLNHPEAVAIITDHILEGARDGRTVAELMISGRDVLGRDDVLV GVPEMLDDVQVEATFPDGTKLVTVHHPIP*
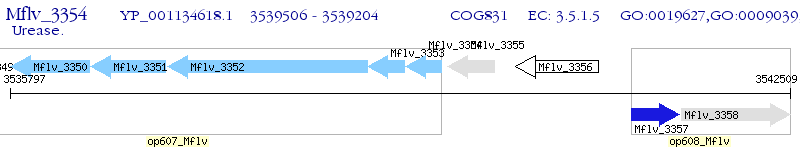
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3354 | ureA | - | 100% (100) | urease subunit gamma |
| M. gilvum PYR-GCK | Mflv_1686 | - | 6e-22 | 53.54% (99) | bifunctional urease subunit gamma/beta |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1879 | ureA | 1e-46 | 87.88% (99) | urease subunit gamma |
| M. tuberculosis H37Rv | Rv1848 | ureA | 1e-46 | 87.88% (99) | urease subunit gamma |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2423 | - | 1e-47 | 87.88% (99) | urease, gamma subunit (UreA) |
| M. marinum M | MMAR_2722 | ureA | 3e-47 | 88.89% (99) | urease gamma subunit UreA |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3627 | ureA | 4e-46 | 86.87% (99) | urease subunit gamma |
| M. thermoresistible (build 8) | TH_1061 | ureA | 2e-48 | 89.00% (100) | Urease gamma subunit ureA (Urea amidohydrolase) |
| M. ulcerans Agy99 | MUL_3031 | ureA | 2e-47 | 88.89% (99) | urease subunit gamma |
| M. vanbaalenii PYR-1 | Mvan_3084 | ureA | 3e-49 | 93.00% (100) | urease subunit gamma |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3354|M.gilvum_PYR-GCK ---------------------MRLTPHEQERLLLSYAAELARRRQARGLK
Mvan_3084|M.vanbaalenii_PYR-1 ---------------------MRLTPHEQERLLLSYAAELARRRQARGLK
MSMEG_3627|M.smegmatis_MC2_155 ---------------------MRLTPHEQDRLLISYAAELARRRRARGLR
TH_1061|M.thermoresistible__bu ---------------VAGSLSVRLTPHEQERLLISYAAELARRRRARGLK
MAB_2423|M.abscessus_ATCC_1997 ---------------------MRLTPHEQERLLISYAAELARRRRSRGLL
Mb1879|M.bovis_AF2122/97 ---------------------MRLTPHEQERLLLSYAAELARRRRARGLR
Rv1848|M.tuberculosis_H37Rv ---------------------MRLTPHEQERLLLSYAAELARRRRARGLR
MMAR_2722|M.marinum_M MTSVTLASKGQNRHGVAGSQSMRLTPHEQERLLLSYAAEVARRRQARGLR
MUL_3031|M.ulcerans_Agy99 ---------------------MRLTPHEQERLLLSYAAEVARRRQARGLR
:*******:***:*****:****::***
Mflv_3354|M.gilvum_PYR-GCK LNHPEAVAIITDHILEGARDGRTVAELMISGRDVLGRDDVLVGVPEMLDD
Mvan_3084|M.vanbaalenii_PYR-1 LNHPEAVAVITDHILEGARDGRTVAELMVSGRAVLGRADVMDGVPEMLHD
MSMEG_3627|M.smegmatis_MC2_155 LNHPEAVAVITDHLLEGARDGRTVAELMVSGRSVLTRDDVMDGVPEMLHD
TH_1061|M.thermoresistible__bu LNHPEAVAMITDHLLEGARDGRTVAELMVSGREVLTRDDVMEGVPEMLSD
MAB_2423|M.abscessus_ATCC_1997 LNHPEAIAIITDHLLEGARDGRTVAELMVSGREVLGRDDVMDGVPEMIPD
Mb1879|M.bovis_AF2122/97 LNHPEAIAVIADHILEGARDGRTVAELMASGREVLGRDDVMEGVPEMLAE
Rv1848|M.tuberculosis_H37Rv LNHPEAIAVIADHILEGARDGRTVAELMASGREVLGRDDVMEGVPEMLAE
MMAR_2722|M.marinum_M LNHPEAVALITDHVLEGARDGRTVAELMASGCEVLGRDDVMEGVPEMIAD
MUL_3031|M.ulcerans_Agy99 LNHPEAVALITDHVLEGARDGRTVAELMASGCEVLGRDDVMEGVPEMIAD
******:*:*:**:************** ** ** * **: *****: :
Mflv_3354|M.gilvum_PYR-GCK VQVEATFPDGTKLVTVHHPIP
Mvan_3084|M.vanbaalenii_PYR-1 VQVEATFPDGTKLVTVHHPIP
MSMEG_3627|M.smegmatis_MC2_155 VQVEATFPDGTKLVTVHQPIA
TH_1061|M.thermoresistible__bu VQVEATFPDGTKLVTVHHPIP
MAB_2423|M.abscessus_ATCC_1997 VQVEATFPDGTKLVTVHHPIG
Mb1879|M.bovis_AF2122/97 VQVEATFPDGTKLVTVHQPIA
Rv1848|M.tuberculosis_H37Rv VQVEATFPDGTKLVTVHQPIA
MMAR_2722|M.marinum_M VQVEATFPDGTKLVTVHHPIR
MUL_3031|M.ulcerans_Agy99 VQVEATFPDGTKLVTVHHPIR
*****************:**