For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DTNVIWIIVAVVVALIVVGVLVALARKSRNRRRAAEADRIREEVHHDTKRVERQATLVEETEAKARAAKA EADAKAAEAEAKAAEAARLADTASTSRDALTSSQEHIDARREHADKLDPRRRTADQADPAVQNREDVNTQ TGVTTHRDDTTHTVGPDTRTR*
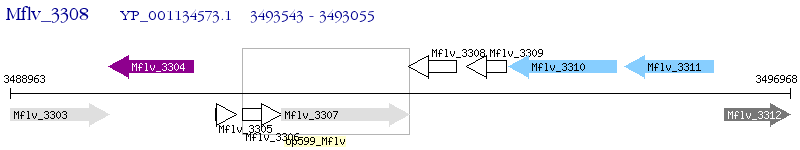
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3308 | - | - | 100% (162) | hypothetical protein Mflv_3308 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_5061 | - | 4e-20 | 38.06% (134) | hypothetical protein MMAR_5061 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3328 | - | 2e-16 | 31.85% (135) | hypothetical protein MSMEG_3328 |
| M. thermoresistible (build 8) | TH_2530 | - | 1e-15 | 42.71% (96) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_4137 | - | 3e-20 | 38.06% (134) | hypothetical protein MUL_4137 |
| M. vanbaalenii PYR-1 | Mvan_3031 | - | 1e-47 | 60.71% (168) | hypothetical protein Mvan_3031 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3308|M.gilvum_PYR-GCK MDTN-VIWIIVAVVVALIVVGVLVALARKSRNRRRAAEADRIREEVHHDT
Mvan_3031|M.vanbaalenii_PYR-1 MDTNNVVWIVIAAVVAIIVIGVIVFLARRARNQRRAQEADRIREEVHHDS
MMAR_5061|M.marinum_M -MNSSMIWIVVGVVAAIVVLGGLIVVASRTRRHRRQQQAEEIREQARIQA
MUL_4137|M.ulcerans_Agy99 -MNSSMIWIVVGVVAAIVVLGGLIVVASRTRRHRRQQQAEEIREQARLQA
MSMEG_3328|M.smegmatis_MC2_155 MPTSSIVLIVVIVIAAILLIAAIVWVARNKRNEHRHVAAEEIREAARDET
TH_2530|M.thermoresistible__bu VDSNTVIMIVVAVVAVLIVVALVVLLARNAQHKRRRVEGERLRREVEEKS
.. :: *:: .:..::::. :: :* . :..:* .:.:*. .. .:
Mflv_3308|M.gilvum_PYR-GCK KRVERQATLVEETEAKARAAKAEADAKAAEAEAKAAEAARLADTASTSRD
Mvan_3031|M.vanbaalenii_PYR-1 RRVQRQASIVEETEAKARAAQAEA-------EAKAAEAARLAETASSRRD
MMAR_5061|M.marinum_M AQLERRQALADETAAKARAAQAEA-------EVKAAEAARLQERAAQHQS
MUL_4137|M.ulcerans_Agy99 AQLERRRALADETAAKARAAQAEA-------EVKAAEAARLQERAAQHQS
MSMEG_3328|M.smegmatis_MC2_155 PQVRQREARADETAARARAAQAES-------NVKAAQASGLQQQAVAYRS
TH_2530|M.thermoresistible__bu RHVEKRESVAAETEARARAAEAEAEX-----XXRGAAGPPVKSTKPAGTV
::.:: : . ** *:****:**: :.* .. : .
Mflv_3308|M.gilvum_PYR-GCK ALTSSQEHIDARREHADKLDPRRRTADQADPAVQNREDVNTQTGVTTHRD
Mvan_3031|M.vanbaalenii_PYR-1 ALNSSQQHLDSRLEHADTLDPRRTDDDKGGAVAPDPKNP-VRDPENPRRD
MMAR_5061|M.marinum_M EAEATRTQLDEQWQRAEDLAPEAGAQSSQTASQSDAVDTGSSR-------
MUL_4137|M.ulcerans_Agy99 EAEATRTQLDEQWQRAEDLAPEAGAQSSQTASQSDAVDTGSSR-------
MSMEG_3328|M.smegmatis_MC2_155 EAETSRHQLDEQWDRADTMDPTS--QTPDMPRTADRKEHQAH--------
TH_2530|M.thermoresistible__bu SGPSTGTQIQRGVTRPGLAGSLSAGDDDAGSDRAAGTQ------------
:: ::: :. . . :
Mflv_3308|M.gilvum_PYR-GCK DTTHTVGPDTRTR--
Mvan_3031|M.vanbaalenii_PYR-1 GTAETMRPDVRTRSH
MMAR_5061|M.marinum_M ---------------
MUL_4137|M.ulcerans_Agy99 ---------------
MSMEG_3328|M.smegmatis_MC2_155 ---------------
TH_2530|M.thermoresistible__bu ---------------