For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRHRRQQIRTHSGIVLRVAAALASVAMVTGMTVAASTTATAAVDTGVATVSPGPGQTVGVAHPVTVTFTE PVRDRTAAENSISVAPAGTSAPMPGTTRWLDGRSVQWEPAEFYPAHTPITVSVGGFSTGFQTGSSVVGVA DLSAYTFTVSIDGVVAREMPASMGKPKFPTPTGSFTALSKERNVTFDSRTIGIPLDDEEGYLIKGEYGVR VTWGGVYVHSAPWSVGSQGYANVSHGCINLSPDNAAWYFDTVSVGDPIIVQA*
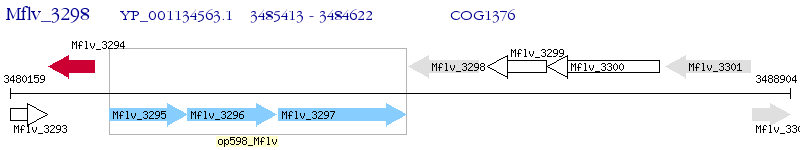
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3298 | - | - | 100% (263) | ErfK/YbiS/YcfS/YnhG family protein |
| M. gilvum PYR-GCK | Mflv_1397 | - | 2e-56 | 45.89% (231) | ErfK/YbiS/YcfS/YnhG family protein |
| M. gilvum PYR-GCK | Mflv_2824 | - | 1e-50 | 39.91% (213) | ErfK/YbiS/YcfS/YnhG family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0120c | - | 1e-76 | 58.40% (250) | hypothetical protein Mb0120c |
| M. tuberculosis H37Rv | Rv0116c | - | 1e-76 | 58.40% (250) | hypothetical protein Rv0116c |
| M. leprae Br4923 | MLBr_02664 | - | 1e-76 | 57.89% (247) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_3165c | - | 9e-79 | 59.44% (249) | hypothetical protein MAB_3165c |
| M. marinum M | MMAR_0316 | - | 3e-78 | 60.58% (241) | hypothetical protein MMAR_0316 |
| M. avium 104 | MAV_5194 | - | 2e-71 | 55.33% (244) | ErfK/YbiS/YcfS/YnhG family protein |
| M. smegmatis MC2 155 | MSMEG_3528 | - | 2e-85 | 64.05% (242) | ErfK/YbiS/YcfS/YnhG family protein |
| M. thermoresistible (build 8) | TH_3496 | - | 4e-81 | 61.09% (239) | PUTATIVE ErfK/YbiS/YcfS/YnhG family protein |
| M. ulcerans Agy99 | MUL_4806 | - | 2e-77 | 60.17% (241) | hypothetical protein MUL_4806 |
| M. vanbaalenii PYR-1 | Mvan_3019 | - | 1e-107 | 71.43% (266) | ErfK/YbiS/YcfS/YnhG family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3298|M.gilvum_PYR-GCK MLRHRRQQIRTHSGIVLRVAAALASVAMVTGMTVAAST---TATAAVDTG
Mvan_3019|M.vanbaalenii_PYR-1 MLRARLQQTRSARGGTRRIAAVLGCAAVVTGVSLAGPVPIGSAGSAVGAA
MSMEG_3528|M.smegmatis_MC2_155 MLNSLGRGSRSGRGSVWKLFAAVGLAVAATAVPVNVAS----AAVSTPVE
TH_3496|M.thermoresistible__bu -----------VREAIRYVCTVLGVVTVLLAGSAGTGT----AAIHNPVP
MAB_3165c|M.abscessus_ATCC_199 ------------------MLVVTGLVSLLMNAAVTAPATLGHALAASPSG
Mb0120c|M.bovis_AF2122/97 -----------MRRVVRYLSVVVAITLMLTAESVSIATAAVPPLQPIP-G
Rv0116c|M.tuberculosis_H37Rv -----------MRRVVRYLSVVVAITLMLTAESVSIATAAVPPLQPIP-G
MMAR_0316|M.marinum_M -----------MRRIIRYLFVMVAITVLAVVAAGSPATAALPAAEPIP-G
MUL_4806|M.ulcerans_Agy99 ---------------------MVAITVLAVVAAGSPATAALPAAEPIP-G
MLBr_02664|M.leprae_Br4923 -----------MYQAVRYLLVMAAIILMAVAESGSPSVAAIPALKPTP-E
MAV_5194|M.avium_104 ---------------------MVAITVMALVAPVGRGGASVPLPQPVP-G
. .
Mflv_3298|M.gilvum_PYR-GCK VATVSPGPGQTVGVAHPVTVTFTEPVRDRTAAENSISVAPAGTSAPMPGT
Mvan_3019|M.vanbaalenii_PYR-1 VASVSPAPGETVGVAHPVTVTFAGQVPDRAAAERAVNVVPAGASAPAAGS
MSMEG_3528|M.smegmatis_MC2_155 VAQVEPAAGAVVGVAHPVTVRFAEPVTDRRSAERSLRIASTDTSA---GR
TH_3496|M.thermoresistible__bu VADIAPADGAIVGVAHPVEVTFAAPVVNRLAAQRSIRIISDNAPA---GE
MAB_3165c|M.abscessus_ATCC_199 VASVSPTPGQTVGVAMPVTIRFAAPVADRIAAERSIEFSAPKVPA---GA
Mb0120c|M.bovis_AF2122/97 VASVSPANGAVVGVAHPVVVTFTTPVTDRRAVERSIRISTPHNTT---GH
Rv0116c|M.tuberculosis_H37Rv VASVSPANGAVVGVAHPVVVTFTTPVTDRRAVERSIRISTPHNTT---GH
MMAR_0316|M.marinum_M VASVLPANGAVVGVAHPVVVTFTAPVTNRRAAQRTIHVLSPSNMT---GH
MUL_4806|M.ulcerans_Agy99 VASVLPANRAVVGVAHPVVVTFTAPVTNRRAAQRTIHVLSPSNMT---GH
MLBr_02664|M.leprae_Br4923 VASVLPTNGAVVGVAHLVVVTFTAPVTDRSAAERSIRITSPNNMT---GH
MAV_5194|M.avium_104 IASILPANGAVVGVAHPIVVTFTAPAADRAAVERSIHVVSPAGVR---GH
:* : * **** : : *: . :* :.:.:: . . *
Mflv_3298|M.gilvum_PYR-GCK TRWLDGR-SVQWEPAEFYPAHTPITVSVG---GFSTGFQTGSSVVGVADL
Mvan_3019|M.vanbaalenii_PYR-1 FTWLDDR-VVEWTPAQPLPAHTAIGVSVG---AFATGFETGASVVSVADI
MSMEG_3528|M.smegmatis_MC2_155 FRWPEAA-VMEWTPDEFWPAHSTISLSVG---GVKTSFNTGAEVLGVADI
TH_3496|M.thermoresistible__bu FSWLGDR-VVRWTPDGYWPAHSSVSVLAG---GIKTNFRTGSVVKGVADI
MAB_3165c|M.abscessus_ATCC_199 FSWVDNA-TVRFTPREYWPAHSSITVSVNGVSGMKYKFQTGSEVLGIGSI
Mb0120c|M.bovis_AF2122/97 FEWVASN-VVRWVPHRYWPPHTRVSVGVQ---ELTEGFETGDALIGVASI
Rv0116c|M.tuberculosis_H37Rv FEWVASN-VVRWVPHRYWPPHTRVSVGVQ---ELTEGFETGDALIGVASI
MMAR_0316|M.marinum_M FQWVQSN-VVQWVPDRYWPTHTRVSVGVQ---ELTTGFETGDELLGVASI
MUL_4806|M.ulcerans_Agy99 FQWVQSN-VVQWVPDRYWPTHTRVSVGVQ---ELTTGFETGDELLGVASI
MLBr_02664|M.leprae_Br4923 FEWLDGD-VVQWIPTKYWPAYTHVSVEVQ---ALTTGFETGDALLGVASL
MAV_5194|M.avium_104 FEWADDNTVVRFVPNRYWPAHGHVSVGVQ---ALTTGFDTGDALLGVASI
* :.: * *.: : : . . * ** : .:..:
Mflv_3298|M.gilvum_PYR-GCK SAYTFTVSIDGVVAREMPASMGKPKFPTPTGSFTALSKERNVTFDSRTIG
Mvan_3019|M.vanbaalenii_PYR-1 SEHTFTVSIDGVVARQMPASMGKPRYPTPTGQFTALEKQRYVTFDSRTIG
MSMEG_3528|M.smegmatis_MC2_155 DAHTFTVSVDGEVLRKMPASMGKPKFPTPRGTFTALAKEPVVVMDSRTIG
TH_3496|M.thermoresistible__bu SAHTFTVSIDGEVVREMPASMGKPKFPTPTGSFSVLGKEKMVVMDSRTIG
MAB_3165c|M.abscessus_ATCC_199 SGHTFTVKIDGTVMRTMPASMGKPKHPTPVGSFTALEKQSPVVMDSRTIG
Mb0120c|M.bovis_AF2122/97 SAHTFTVSRNGEVLRTMPASLGKPSRPTPIGSFHAMSKERTVVMDSRTIG
Rv0116c|M.tuberculosis_H37Rv SAHTFTVSRNGEVLRTMPASLGKPSRPTPIGSFHAMSKERTVVMDSRTIG
MMAR_0316|M.marinum_M SAHTFTVSRNGDVLRTMPASMGKPSRPTPIGSFTALSKERTVVMDSRTIG
MUL_4806|M.ulcerans_Agy99 SAHTFTVSRNGDVLRTMPASMGKPSRPTPIGSFTALSKERTVVMDSRTIG
MLBr_02664|M.leprae_Br4923 STHTFTVSRNGEVLRTMPASMGKPTRPTPIGKFTALSKERTVVMDSRTIG
MAV_5194|M.avium_104 SKHTFTVSRNGEVLRTMPASMGKPSRPTPIGNFTALEKQRSVVMDSRTIG
. :****. :* * * ****:*** *** * * .: *: *.:******
Mflv_3298|M.gilvum_PYR-GCK IPLDDEEGYLIKGEYGVRVTWGGVYVHSAPWSVGSQGYANVSHGCINLSP
Mvan_3019|M.vanbaalenii_PYR-1 IPLDDPEGYLIKGEYGVRVTWGGVYVHSAPWSVGSQGYANVSHGCINLSP
MSMEG_3528|M.smegmatis_MC2_155 IPLSDPEGYKLTVNHAVRVTWGGVYVHSAPWSVGSQGYANVSHGCINLSP
TH_3496|M.thermoresistible__bu IPLDDPEGYRLNVENAVRITWGGVYIHSAPWSVGSQGYANVSHGCINLSP
MAB_3165c|M.abscessus_ATCC_199 IPLNDPEGYKLTVYYAVRVTWGGVYVHSAPWSTGAQGNSNVSHGCINLSP
Mb0120c|M.bovis_AF2122/97 IPLNSSDGYLLTAHYAVRVTWSGVYVHSAPWSVNSQGYANVSHGCINLSP
Rv0116c|M.tuberculosis_H37Rv IPLNSSDGYLLTAHYAVRVTWSGVYVHSAPWSVNSQGYANVSHGCINLSP
MMAR_0316|M.marinum_M IPLNSSEGYMITASYAVRVTWSGVYVHSAPWSVNSQGYANVSHGCINLSP
MUL_4806|M.ulcerans_Agy99 IPLNSSEGYMITASYAVRVTWSGVYVHSAPWSVNSQGYANVSHGCINLSP
MLBr_02664|M.leprae_Br4923 IPLNSPEGYLITAQYAVRVTWSGVYVHSAPWSVNSQGYTNVSHGCINLSP
MAV_5194|M.avium_104 IPLSSPEGYKITAQYAVRVTWSGVYVHSAPWSVDSQGYANVSHGCINLSP
***.. :** :. .**:**.***:******..:** :***********
Mflv_3298|M.gilvum_PYR-GCK DNAAWYFDTVSVGDPIIVQA
Mvan_3019|M.vanbaalenii_PYR-1 DNAAWYFDTVGVGDPIIVQA
MSMEG_3528|M.smegmatis_MC2_155 DNAAWYYDMVSVGDPIIVQA
TH_3496|M.thermoresistible__bu SNAAWYFDQVRIGDPVIVQP
MAB_3165c|M.abscessus_ATCC_199 DNASWYYNTVSIGDPIIINA
Mb0120c|M.bovis_AF2122/97 DNAAWYFDAVTVGDPIEVVG
Rv0116c|M.tuberculosis_H37Rv DNAAWYFDAVTVGDPIEVVG
MMAR_0316|M.marinum_M DNAAWYFDSVNVGDPIEVVG
MUL_4806|M.ulcerans_Agy99 DNAAWYFDSVNVGDPIEVVG
MLBr_02664|M.leprae_Br4923 DDATWYFNTVNVGDPIEVVA
MAV_5194|M.avium_104 DNAAWYFNEVNVGDPIQVVA
.:*:**:: * :***: :