For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALVSPISQLEERRQPTTTLEGWRRFIDADRPEFTLLPDQDWSDLDDADRIAYNEARVAHHSELVVVTTSA IQKITNEGQLLTLLNQREIGARRGLIVSGGAATGKTTAIKQLGRFHELRIRTRYSDKTRIPVVYVTAPPK GSARKLAMEFARFLGLPPVRRMNVTDIADAVCQVLIDARTDIVVVDEIHNLNLDTRAGEELSDHLKYFTE HLPATFVYAGIDVERSGVFTGTRGRQLAGRCGVINTAAFPYGQEWRALVAAMEGTLRLHRHEPGTLVRQT KHLHQRTGGMIGSLAHLIRSAAIRAMLDHTEHIDREGLDSVLIDYAAQASAQRSAG*
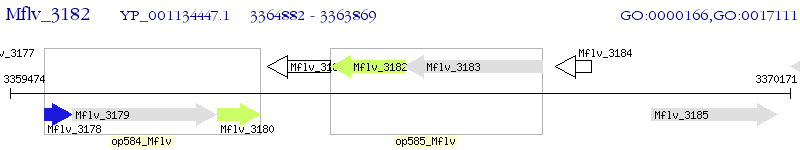
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3182 | - | - | 100% (337) | putative ATP/GTP-binding protein |
| M. gilvum PYR-GCK | Mflv_0612 | - | 0.0 | 100.00% (337) | ATP/GTP-binding protein |
| M. gilvum PYR-GCK | Mflv_3219 | - | e-156 | 80.18% (338) | ATP/GTP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4892c | - | 2e-29 | 30.40% (329) | hypothetical protein MAB_4892c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3321 | - | 1e-151 | 80.72% (332) | ATP/GTP-binding protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3182|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3321|M.smegmatis_MC2_155 MQPISHSTLVMAATESSQSSQSSQSSHFAASARGVRRRSAGTLPELEDLL
MAB_4892c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3182|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3321|M.smegmatis_MC2_155 TALRLDDAFTGDAVITGDDPLINSPHRLTTASATALLLGGAAAAAVWSAR
MAB_4892c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3182|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3321|M.smegmatis_MC2_155 TGQQTDVAIDAVHALHHLHPTHFVTQQGHPMNVGAEYVAANGVFATRDGH
MAB_4892c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3182|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3321|M.smegmatis_MC2_155 HIMIEAGPPYQKLLNGYLEFFDCPNTKEALQHRIRQWDAEELETALAAAG
MAB_4892c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3182|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3321|M.smegmatis_MC2_155 LPGCRAFTPAQWRKHPQGALLARTPVITIDKIADGDPVPFSTTPTAPLTG
MAB_4892c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3182|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3321|M.smegmatis_MC2_155 TRVLDFTHVLAGPRSTQTLAEYGADVLHVSSPRYPDTLAQHLCVDRGKRC
MAB_4892c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3182|M.gilvum_PYR-GCK -------------------------------MALVSP-----ISQLEERR
MSMEG_3321|M.smegmatis_MC2_155 TYLDLARESDAERCGRGDSSAGVRCTQGGPIVATVTEPQTVAVNQLEDRR
MAB_4892c|M.abscessus_ATCC_199 ----------------------------------MSP-----QPRRNLQN
:: : : :.
Mflv_3182|M.gilvum_PYR-GCK QPTTTLEGWRRFIDAD-RPEFTLLPDQDWSDLDDADRIAYNEARVAHHSE
MSMEG_3321|M.smegmatis_MC2_155 QPTTTLEGWRRFIEAD-APEFELLDDLTWSQLDDDARTRYNEARVAHHSE
MAB_4892c|M.abscessus_ATCC_199 ITLIRKEGWRDFVVAAPRRQPEQLTTAELAVLSPRAAAQYNTRRADWHAN
. **** *: * : * : *. ** *. *::
Mflv_3182|M.gilvum_PYR-GCK LVVVTTSAIQKITNEGQLLTLLNQREIG-ARRGLIVSGGAATGKTTAIKQ
MSMEG_3321|M.smegmatis_MC2_155 LVVVTTSAIESITNQGRLLTLLNQREIG-ARRGLIISGAAATGKTTAIKQ
MAB_4892c|M.abscessus_ATCC_199 MGVLKTHQLKAVQEMLDLILESNQQDGDKAKDAIALSASAGLGKTTAVED
: *:.* :: : : *: **:: . *: .: :*..*. *****:::
Mflv_3182|M.gilvum_PYR-GCK LGRFHELR----IRTRYS-DKTRIPVVYVTAPPKGSARKLAMEFARFLGL
MSMEG_3321|M.smegmatis_MC2_155 LGRFHELR----TRARFPGDDNRIPVVYVTAPPKGSPRKLAMEFARFLGL
MAB_4892c|M.abscessus_ATCC_199 FGKKYHLSQIQTHGPRTTGGHERWPVCRITLTGSTTIRDFNRALLEFFAH
:*: :.* .* . .. * ** :* . . : *.: : .*:.
Mflv_3182|M.gilvum_PYR-GCK PPVR-RMNVTDIADAVCQVLIDARTDIVVVDEIHNLNLDTRAGEELSDHL
MSMEG_3321|M.smegmatis_MC2_155 PMLNPRMNVTDISDAVCQVLIDARTDIVVVDEIHNLNLDTRAGEELSDHL
MAB_4892c|M.abscessus_ATCC_199 PGAD-RGTAAQYLRRALDCFLSCQVRLLIVDDVHFLQPRSRNGVEVSNHF
* * ..:: . : ::..:. :::**::* *: :* * *:*:*:
Mflv_3182|M.gilvum_PYR-GCK KYFTEHLPATFVYAGIDVERSGVFTGTRGRQLAG---RCGVINTAAFPYG
MSMEG_3321|M.smegmatis_MC2_155 KYFTEHLPATFVYAGIDVERSGLFTGTRGRQLAG---RCGVIHTSAFPDA
MAB_4892c|M.abscessus_ATCC_199 KFITSEFPVTLLLVGIGLENIGLFPRGVDHTLAQTGRRTTILTMDPYTDT
*::*..:*.*:: .**.:*. *:*. .: ** * :: .:.
Mflv_3182|M.gilvum_PYR-GCK -QEWRALVAAMEGTLRLHRHEPGTLVR-QTKHLHQRTGGMIGSLAHLIRS
MSMEG_3321|M.smegmatis_MC2_155 -KEWRQLVAAMEGTLRLHSHPPGSLVA-EARYLHRRTGGMIGSLAHLIRA
MAB_4892c|M.abscessus_ATCC_199 GEQWRALLLAIEQRLVLANKHQGMLADTLSDYVYRRTGGHIGSLMTLINR
::** *: *:* * * : * *. : ::::**** **** **.
Mflv_3182|M.gilvum_PYR-GCK AAIRAMLDHTEHIDREGLDSVLIDYAAQASAQRSAG------------
MSMEG_3321|M.smegmatis_MC2_155 AAIQAMLDGTEQVSRALMDTVLIDYAAHTAAARTAS------------
MAB_4892c|M.abscessus_ATCC_199 GSLHAIRTGTEALTEELLDAVPIDFAAEGQRGAREAAHRNSTRRLRRQ
.:::*: ** : . :*:* **:**. .