For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLRSYCDFYEVDPPGDKLDAMCRALIADPSEGVQLIARDASGETVGFATIFWTWQTLYAARVGVLNDLYV TPRSRGAGTGRALIARGMQMCHERGAEKLVWETSPDNATAQRLYDGIGAEKSVWLTYELGT
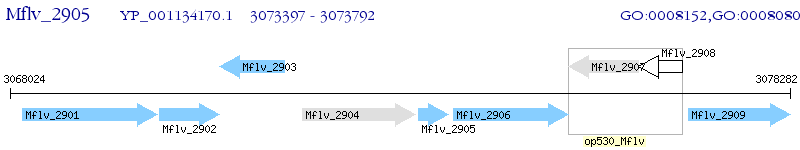
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2905 | - | - | 100% (131) | GCN5-related N-acetyltransferase |
| M. gilvum PYR-GCK | Mflv_4681 | - | 1e-07 | 30.43% (92) | GCN5-related N-acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3242 | - | 5e-05 | 28.26% (92) | acetyltransferase |
| M. tuberculosis H37Rv | Rv3216 | - | 5e-05 | 28.26% (92) | acetyltransferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3536 | - | 2e-09 | 34.78% (92) | putative acetyltransferase |
| M. marinum M | MMAR_1341 | - | 3e-07 | 31.52% (92) | acetyltransferase |
| M. avium 104 | MAV_4165 | - | 5e-09 | 30.77% (130) | acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_1924 | - | 2e-06 | 29.35% (92) | N-acetyltransferase Ats1 |
| M. thermoresistible (build 8) | TH_0671 | - | 1e-06 | 32.71% (107) | POSSIBLE ACETYLTRANSFERASE |
| M. ulcerans Agy99 | MUL_2538 | - | 2e-07 | 31.52% (92) | acetyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1786 | - | 9e-08 | 31.52% (92) | GCN5-related N-acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1924|M.smegmatis_MC2_155 MT-ELIRRARPGDEVEITAMIRELAEFEHASDECTVTEEQLHTALFGDKP
Mvan_1786|M.vanbaalenii_PYR-1 MS-EIIRAVRPGDEAELTAMIHELAKFEHAAAECTVTENQLAAALFGPDP
TH_0671|M.thermoresistible__bu VT-VQIRPARLGDEVELAAMVHELAEFEHAAAECTVTPAQLRTALFGDPV
Mb3242|M.bovis_AF2122/97 --------------------------------------------------
Rv3216|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1341|M.marinum_M MT-ENIRRAKPEDTAEITAMIRELAEFERAADQCTVSEKQISTALFGALP
MUL_2538|M.ulcerans_Agy99 MT-ENIRRAKPEDTAEITAMIRELAEFERAADQCTVSEKQIFTALFGALP
MAV_4165|M.avium_104 MS-HRIRRAAPHDAAAITEMIHALAEFERAADQCTVTETRLAAALFDASP
MAB_3536|M.abscessus_ATCC_1997 MTNTLIRRATEADIPAIVGLIEDLAEYEHARDECLITEEQLHQALFGSHP
Mflv_2905|M.gilvum_PYR-GCK -------------------MLRSYCDFYEVDPPGDKLDAMCRALIADPSE
MSMEG_1924|M.smegmatis_MC2_155 VAYAHVVEVDGQAAATAVWFLNFSTWDGVAGIYLEDLFVRPAFRRRGIAR
Mvan_1786|M.vanbaalenii_PYR-1 TLYGHLVEVDGQAAATALWFRSFSTWDGVAGVYLEDLFVRPQFRRRGLAR
TH_0671|M.thermoresistible__bu TVRGHLAEVDGEVAACALWFHNFSTWDGVAGIHLEDLFVRPRFRRRGVAR
Mb3242|M.bovis_AF2122/97 -MRGHVAEVNGGVAAMALWFLNFSTWDGVAGIYVEDLFVWPRFRRRGLAR
Rv3216|M.tuberculosis_H37Rv -MRGHVAEVNGGVAAMALWFLNFSTWDGVAGIYVEDLFVWPRFRRRGLAR
MMAR_1341|M.marinum_M TVRGHIVEIDGQIAAMALWFLNFSTWDGAAGIYLEDLYVRPQFRRRGLAR
MUL_2538|M.ulcerans_Agy99 TVRGHIVEIDGQIAAMALWFLNFSTWDGAAGIYLEDLYVRPQFRRRGLAR
MAV_4165|M.avium_104 TVHGHVAEVDGTIAGMALWFVNFSTWDGVAGIYLEDLFVRPGFRRRGLAR
MAB_3536|M.abscessus_ATCC_1997 ALFAHVAESGGVVCGTAVWFLNFSTWTGHHGIYLEDLYVRPEQRGAGLGK
Mflv_2905|M.gilvum_PYR-GCK GVQLIARDASGETVGFATIFWTWQTLYAARVGVLNDLYVTPRSRGAGTGR
: .* . * * .:.* . ::**:* * * * .:
MSMEG_1924|M.smegmatis_MC2_155 KLLATLARECVDNGYSRLQWAVLNWNVNAIALYDAVGGKPQTQWTTYRVS
Mvan_1786|M.vanbaalenii_PYR-1 KLLSTLARECVERGYSRLSWAVLDWNVNAIALYDSVGGTPQSEWITYRVS
TH_0671|M.thermoresistible__bu ALLATLARECVANGYTRLSWAVLDWNRDAIALYESVRAKPLDEWTTYRLS
Mb3242|M.bovis_AF2122/97 GLLSTLARECVDNRYTRLAWSVLNWNSDAIALYDRIGGQPQHEWTIYRLS
Rv3216|M.tuberculosis_H37Rv GLLSTLARECVDNRYTRLAWSVLNWNSDAIALYDRIGGQPQHEWTIYRLS
MMAR_1341|M.marinum_M RLLATLAAECLDNGYTRLSWAVLNWNSNAIAVYDAVGGQPQNEWTTYRLS
MUL_2538|M.ulcerans_Agy99 RLLATLAAECLDNGYTRLSWAVLNWNSNAIAVYDAVGGQPQNEWTTYRLS
MAV_4165|M.avium_104 ALLAALAAECVRRGYTRLSWAVLNWNTDAIALYDGIGARPQREWTTYRLS
MAB_3536|M.abscessus_ATCC_1997 ALLSALAAECVANGYTRLDWAVLDWNTPSIKFYDSLGAHPQSDWITYRLQ
Mflv_2905|M.gilvum_PYR-GCK ALIARGMQMCHERGAEKLVWETSPDNATAQRLYDGIG-AEKSVWLTYELG
*:: * . :* * . * : .*: : * *.:
MSMEG_1924|M.smegmatis_MC2_155 GRELTALAAE---
Mvan_1786|M.vanbaalenii_PYR-1 GPRLSELAAESRD
TH_0671|M.thermoresistible__bu GPALSALAES---
Mb3242|M.bovis_AF2122/97 GPRLAALAAPR--
Rv3216|M.tuberculosis_H37Rv GPRLAALAAPR--
MMAR_1341|M.marinum_M GPDLVALAGPR--
MUL_2538|M.ulcerans_Agy99 GPDLVALAGPR--
MAV_4165|M.avium_104 GPRLAELAESS--
MAB_3536|M.abscessus_ATCC_1997 GPALAELASDRSD
Mflv_2905|M.gilvum_PYR-GCK T------------