For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAVRLADVIDVLDAAYPPRLAQSWDSVGLVCGDPSEPVESVTIAVDATADVLAQVPQGGLLLAHHPLLLR GVDTVAADTAKGALIHSAIRTGRSLFTAHTNADSASPGVSDALAETLGLEVCEVLSPVPSGPSLDKWAVF VPAENAEAVRTAMFAAGAGHIGDYSQCSWSVSGTGQFLPGEGASPAVGAVGAVERLAEDRVEMVAPTSRR AAVLAGLRAAHPYEEPAFDLFPLQTPPGDVGLGRIATLREAEPLAAFVSRVRAALPATSWGVRAAGDPAA TVSRVAVCGGAGDSLLAEVAGAGVQAYVTADLRHHPADEHRRVSDVALIDVAHWASEFPWCAQAAELLRG QFGPALPVTVSDVRTDPWNVEGC
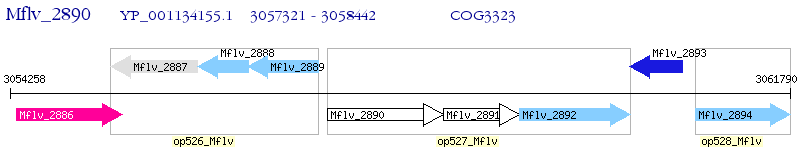
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2890 | - | - | 100% (373) | hypothetical protein Mflv_2890 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2255c | - | 1e-158 | 73.85% (371) | hypothetical protein Mb2255c |
| M. tuberculosis H37Rv | Rv2230c | - | 1e-158 | 73.85% (371) | hypothetical protein Rv2230c |
| M. leprae Br4923 | MLBr_01639 | - | 1e-154 | 71.35% (370) | hypothetical protein MLBr_01639 |
| M. abscessus ATCC 19977 | MAB_1903 | - | 1e-137 | 64.97% (374) | hypothetical protein MAB_1903 |
| M. marinum M | MMAR_3306 | - | 1e-159 | 74.32% (370) | hypothetical protein MMAR_3306 |
| M. avium 104 | MAV_2209 | - | 1e-166 | 77.63% (371) | hypothetical protein MAV_2209 |
| M. smegmatis MC2 155 | MSMEG_4307 | - | 1e-171 | 79.40% (369) | hypothetical protein MSMEG_4307 |
| M. thermoresistible (build 8) | TH_1465 | - | 1e-169 | 78.38% (370) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1322 | - | 1e-159 | 74.05% (370) | hypothetical protein MUL_1322 |
| M. vanbaalenii PYR-1 | Mvan_3620 | - | 0.0 | 84.37% (371) | hypothetical protein Mvan_3620 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2890|M.gilvum_PYR-GCK -----MAVRLADVIDVLDAAYPPRLAQSWDSVGLVCGDPSEPVESVTIAV
Mvan_3620|M.vanbaalenii_PYR-1 ----MATVRLVDVIDVLEAAYPPGLAQSWDSVGLVCGDPTEPVESVTIAV
MSMEG_4307|M.smegmatis_MC2_155 MNTPSAGVRLADVIAVLEAAYPPQLAQDWDSVGLVCGDPDEPVESVTIAV
TH_1465|M.thermoresistible__bu -----VTIRLADIIEVLDEAYPPQLAQDWDSVGLVCGDPSEPVESVTVAV
MMAR_3306|M.marinum_M -----MSVRLADVIEVLDGAYPPRLAESWDSVGLVCGDPGEPVESVTVAV
MUL_1322|M.ulcerans_Agy99 -----MSVRLADVIEVLDGAYPPRLAESWDSVGLVCGDPGEPVESVTVAV
Mb2255c|M.bovis_AF2122/97 -----MSVRLADVIDVLDQAYPPRLAQSWDSVGLVCGDPDDVVDSVTVAV
Rv2230c|M.tuberculosis_H37Rv -----MSVRLADVIDVLDQAYPPRLAQSWDSVGLVCGDPDDVVDSVTVAV
MLBr_01639|M.leprae_Br4923 -----MSARLADVIEVLDHAYPPRFAQSWDSVGLVCGDPEDVLEAITIAV
MAV_2209|M.avium_104 -----MSVKLADVIAVLDEAYPPGLAEPWDSVGLVCGDPDEPVESVTVAV
MAB_1903|M.abscessus_ATCC_1997 -----MGVRLADIIGVLDAAYPPALAESWDSVGLVCGDPDQQIDTVTVAV
:*.*:* **: **** :*: *********** : ::::*:**
Mflv_2890|M.gilvum_PYR-GCK DATADVLAQV--PQGGLLLAHHPLLLRGVDTVAADTAKGALIHSAIRTGR
Mvan_3620|M.vanbaalenii_PYR-1 DATPQVVASV--PDRALLLVHHPLLLRGVDSVAADTAKGALIHSLIRTGR
MSMEG_4307|M.smegmatis_MC2_155 DATADVIAEV--PDRGLLLAHHPLLLRGVDTVAASTAKGALIHRLIRTGR
TH_1465|M.thermoresistible__bu DATPAVAAEV--PERGLLLAHHPLLLRGVDTVAADTPKGALIHDLIRTGR
MMAR_3306|M.marinum_M DATPAVVDEV--PERGLLLAHHPLLLRGVDTVAANTAKGALIHHLIRTGR
MUL_1322|M.ulcerans_Agy99 DATPAVVDEV--PERGLLLAHHPLLLRGVDTVAANTAKGALIHRLIRTGR
Mb2255c|M.bovis_AF2122/97 DATPAVVDQV--PQAGLLLVHHPLLLRGVDTVAANTPKGVLVHRLIRTGR
Rv2230c|M.tuberculosis_H37Rv DATPAVVDQV--PQAGLLLVHHPLLLRGVDTVAANTPKGVLVHRLIRTGR
MLBr_01639|M.leprae_Br4923 DATPAVIDEV--PDSGLLLVHHPLLLHGVDTVAVSTPKGALVHRLIRSGR
MAV_2209|M.avium_104 DATPAVVDEV--PAGGLLLAHHPLLLRGVDTVAASTPKGALVHRLIRTGR
MAB_1903|M.abscessus_ATCC_1997 DATAAVVDEMDGPGAHLLLAHHPLLLRGVDTVAASTPKGALVHRLIRAGG
***. * .: * ***.******:***:**..*.**.*:* **:*
Mflv_2890|M.gilvum_PYR-GCK SLFTAHTNADSASPGVSDALAETLGLEVCEVLSPVPSGPSLDKWAVFVPA
Mvan_3620|M.vanbaalenii_PYR-1 ALFTAHTNADSASPGVSDALADALGLQVCDVLSPVPSGPALDKWVVFVPA
MSMEG_4307|M.smegmatis_MC2_155 ALFTAHTNADSANPGVSDALAETLGLRVEDVLSPVAAGPNLDKWVVFVPA
TH_1465|M.thermoresistible__bu ALFTAHTNADSADPGVSDALAEALGLTVEEVLSPVPAGPDLDKWVVFVPQ
MMAR_3306|M.marinum_M SLFTAHTNADSASPGVSDALAQALGLTVESVLARSPSVAELDKWVIYVPR
MUL_1322|M.ulcerans_Agy99 SLFTAHTNADSASPGMSDALAQALGLTVESVLARSPSVAELDKWVIYVPR
Mb2255c|M.bovis_AF2122/97 SLFTAHTNADSASPGVSDALAHAVGLTVDAVLDPVPGAADLDKWVIYVPR
Rv2230c|M.tuberculosis_H37Rv SLFTAHTNADSASPGVSDALAHAVGLTVDAVLDPVPGAADLDKWVIYVPR
MLBr_01639|M.leprae_Br4923 SLFTAHTNADSASPGVSDALAHVFGLTVDAVLEPLLGVASLDKWVIYVPL
MAV_2209|M.avium_104 SLFTAHTNADSASPGVSDALADALGLTVEAVLEPARPASDLDKWVIYVPG
MAB_1903|M.abscessus_ATCC_1997 ALFTAHTNADSASPGVSDALAQALGLTVTSVLAP--RFTDYDKWVVFVPT
:***********.**:*****...** * ** . ***.::**
Mflv_2890|M.gilvum_PYR-GCK ENAEAVRTAMFAAGAGHIGDYSQCSWSVSGTGQFLPGEGASPAVGAVGAV
Mvan_3620|M.vanbaalenii_PYR-1 GNAEAVRSAMFAGGAGQLGDYSQCSWSVSGTGQFLPGDGATPAIGSVGTL
MSMEG_4307|M.smegmatis_MC2_155 ADADALREALFAAGAGRIGDYSHCSWSVAGTGQFLPHDGASPAIGEVGTV
TH_1465|M.thermoresistible__bu DDAEKVREAMFAAGAGRIGDYSHCSWSVPGTGQFLPHDGAQPAIGSVGTV
MMAR_3306|M.marinum_M EHTEPVQAAIFDAGAGHIGDYSHCSWSVTGIGQFLPLDGASPAIGSVGTV
MUL_1322|M.ulcerans_Agy99 EHTEPVQAAIFDAGAGHIGDYSHCSWSVTGIGQFLPLDGASPAIGSVGTV
Mb2255c|M.bovis_AF2122/97 ENSEAVRAAVFEAGAGHIGDYSHCSWSVAGTGQFLAHDGASPAIGSVGTV
Rv2230c|M.tuberculosis_H37Rv ENSEAVRAAVFEAGAGHIGDYSHCSWSVAGTGQFLAHDGASPAIGSVGTV
MLBr_01639|M.leprae_Br4923 EHVAAVQAAVFEAGAGHIGDYSHCSWSVTGTGQFMPHDGASPVVGSIGAI
MAV_2209|M.avium_104 ENADAVREAVFAAGAGHIGDYSHCSWSVTGIGQFLPHEGASPAVGSVGRV
MAB_1903|M.abscessus_ATCC_1997 ESVHAVRRAMFEAGAGHIGAYSDCSWSVTGTGQFRPLAGSDPAVGEHGVV
:: *:* .***::* **.*****.* *** . *: *.:* * :
Mflv_2890|M.gilvum_PYR-GCK ERLAEDRVEMVAPTSRRAAVLAGLRAAHPYEEPAFDLFPLQTPPGDVGLG
Mvan_3620|M.vanbaalenii_PYR-1 EQVAEDRVEMVAPASRRAAVLAGLRAAHPYEEPAFDVVALQTPPGDVGLG
MSMEG_4307|M.smegmatis_MC2_155 EQVSEDRVEVIAPARLRAKVLAAMRAAHPYEEPAFDILPLAPIPGDVGIG
TH_1465|M.thermoresistible__bu ERVAEDRVEMIAPARVRAAVLAALRSAHPYEEPAFDIFALAPVPGAAGLG
MMAR_3306|M.marinum_M ERVTEDRVEVIAPARARAAVLSAMRAAHPYEEPAFDILALVPPPADVGLG
MUL_1322|M.ulcerans_Agy99 ERVTEDRVEVIAPARARAAVLSAMRAAHPYEEPAFDILALVPPPADVGLG
Mb2255c|M.bovis_AF2122/97 ERVAEDRVEVVAPARARAEVLAAMRAAHPYEEPAFDIFALVPPPVGSGLG
Rv2230c|M.tuberculosis_H37Rv ERVAEDRVEVVAPARARAEVLAAMRAAHPYEEPAFDIFALVPPPVGSGLG
MLBr_01639|M.leprae_Br4923 ERVAEDRVEVVAPARARAAVLSAMHAAHPYEEPAFDIFALVPPPGDVGLG
MAV_2209|M.avium_104 ERVAEERVEVVAPARARAAVLAAMRAAHPYEEPAFDIFALLPPPGDAGLG
MAB_1903|M.abscessus_ATCC_1997 EQVVEDRVEMVAPSRLRCELFAAIQAAHPYEVPAIDVLPMAPAPSGTGLG
*:: *:***::**: *. :::.:::***** **:*:..: . * *:*
Mflv_2890|M.gilvum_PYR-GCK RIATLREAEPLAAFVSRVRAALPATSWGVRAAGDPAATVSRVAVCGGAGD
Mvan_3620|M.vanbaalenii_PYR-1 RIATLPEPEPLPAFVSRVRRALPATSWGVRASGDPATTVARVAVCGGSGD
MSMEG_4307|M.smegmatis_MC2_155 RIASLPEPEPLSAFVRRVRAALPPTSWGVRASGDPDAVVSRVAVCGGAGD
TH_1465|M.thermoresistible__bu RICSLPEPERLAAFVSRVRKALPATSWGVRAAGDPDAPISRVAVCGGAGD
MMAR_3306|M.marinum_M RIGTLDRPQTLRDFVSRVGAVLPQTSWGVRAAGDPEMLVTRVAVCGGAGD
MUL_1322|M.ulcerans_Agy99 RIGTLDRPQTLRDFVSRVGAVLPQTSWGVRAAGDPEMLVTRVAVCGGAGD
Mb2255c|M.bovis_AF2122/97 RIGRLPKPEPLRTFVARLEAALPPTATGVRAAGDPDLLVSRVAVCGGAGD
Rv2230c|M.tuberculosis_H37Rv RIGRLPKPEPLRTFVARLEAALPPTATGVRAAGDPDLLVSRVAVCGGAGD
MLBr_01639|M.leprae_Br4923 RIGTLPRPESLSAFVARVGAALPQTSSGVRATGDPDMLVSRVAVCGGAGD
MAV_2209|M.avium_104 RIARLEYPEPLRRFVSRVDAALPATSWGVRAAGDPELAVSRVAVCGGAGD
MAB_1903|M.abscessus_ATCC_1997 RIGALPTPETLRDFVSRVRGALPETVWGVRAAGDPDELIQTVAVCGGAGD
** * .: * ** *: .** * ****:*** : ******:**
Mflv_2890|M.gilvum_PYR-GCK SLLAEVAGAGVQAYVTADLRHHPADEHRRVSDVALIDVAHWASEFPWCAQ
Mvan_3620|M.vanbaalenii_PYR-1 SLLSEVAASGVQAYVTADLRHHPADEHRRSSDVALIDAAHWATEFPWCNQ
MSMEG_4307|M.smegmatis_MC2_155 SLLGAVTAAGVQAYVTADLRHHPADEHRRASDVALVDVAHWASEFPWCAQ
TH_1465|M.thermoresistible__bu SLLGTVAKAGVQAYVTSDLRHHPADEHRRASPVALVDVAHWASEYPWCEQ
MMAR_3306|M.marinum_M SLLGAAAAADVQAYLTADLRHHPADEHRRASAVALIDVAHWASEFPWCGQ
MUL_1322|M.ulcerans_Agy99 SLLGAAAAADVQAYLTADLRHHPADEHRRASAVALIDVAHWASEFPWCGQ
Mb2255c|M.bovis_AF2122/97 SLLATVAAADVQAYVTADLRHHPADEHCRASQVALIDVAHWASEFPWCGQ
Rv2230c|M.tuberculosis_H37Rv SLLATVAAADVQAYVTADLRHHPADEHCRASQVALIDVAHWASEFPWCGQ
MLBr_01639|M.leprae_Br4923 SLLSLAAVADVQAYVTADLRHHPADEHRRASNVALIDVAHWASEFPWCGQ
MAV_2209|M.avium_104 SLLAAAAGAGVQAYLTADLRHHPADEHRRASEVALIDVAHWASEFPWCAQ
MAB_1903|M.abscessus_ATCC_1997 SLLGAAVKSGADVYVTADLRHHPADEHARTSSVALVDVAHWASEFPWCAQ
***. .. :..:.*:*:********** * * ***:*.****:*:*** *
Mflv_2890|M.gilvum_PYR-GCK AAELLRGQFGPALPVTVSDVRTDPWNVEG-------C------
Mvan_3620|M.vanbaalenii_PYR-1 AAELLRHHFGASLPVTVSDVRTDPWNIEG-------C------
MSMEG_4307|M.smegmatis_MC2_155 AAGVLRKHFSDALPVRVCEVRTDPWNVES-------LEEHES-
TH_1465|M.thermoresistible__bu AAQVLRARFGADLPVRVSTVRTDPWNVAP-------VTERES-
MMAR_3306|M.marinum_M AAEVLRSHFGEALPVRVCPLRTDPWNVAA-------SRENDS-
MUL_1322|M.ulcerans_Agy99 AAEVLRSHFGEALPVRVCPLRTDPWNVAT-------SRENDS-
Mb2255c|M.bovis_AF2122/97 AAEVLRSHFGASLPVRVCTICTDPWNLDH-------ETGRDQA
Rv2230c|M.tuberculosis_H37Rv AAEVLRSHFGASLPVRVCTICTDPWNLDH-------ETGRDQA
MLBr_01639|M.leprae_Br4923 AADVLRSHFGTALSVRVCTIRTDPWNLGARRVNDVSDSGRDQ-
MAV_2209|M.avium_104 AADLLRSRFGGRLDVRVSSIRTDPWNVEH-------DGGEGR-
MAB_1903|M.abscessus_ATCC_1997 AARVLDTEFGENLSTRISPVRTDPWTLGG-------PEKEEKA
** :* .*. * . :. : ****.: