For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IGYVAAAAVGYVLGTKAGRRRFEQISGTYRAVTESPAAKAVLDAGRRKIAERVSPDPAMVKLTRIDSGTE VLEPEPIVKKESREGR*
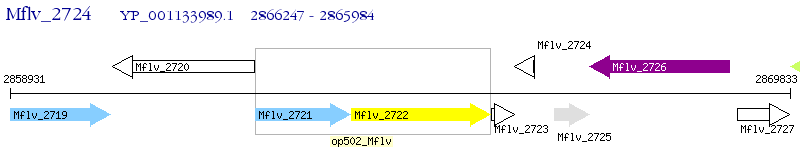
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2724 | - | - | 100% (87) | hypothetical protein Mflv_2724 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2371 | - | 4e-21 | 58.11% (74) | hypothetical protein Mb2371 |
| M. tuberculosis H37Rv | Rv2342 | - | 4e-21 | 58.11% (74) | hypothetical protein Rv2342 |
| M. leprae Br4923 | MLBr_00834 | - | 3e-19 | 55.41% (74) | hypothetical protein MLBr_00834 |
| M. abscessus ATCC 19977 | MAB_1709c | - | 3e-20 | 54.88% (82) | hypothetical protein MAB_1709c |
| M. marinum M | MMAR_3649 | - | 3e-25 | 68.83% (77) | hypothetical protein MMAR_3649 |
| M. avium 104 | MAV_2044 | - | 9e-22 | 59.21% (76) | hypothetical protein MAV_2044 |
| M. smegmatis MC2 155 | MSMEG_4479 | - | 3e-24 | 68.42% (76) | hypothetical protein MSMEG_4479 |
| M. thermoresistible (build 8) | TH_0724 | - | 2e-28 | 75.00% (76) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1460 | - | 2e-25 | 68.83% (77) | hypothetical protein MUL_1460 |
| M. vanbaalenii PYR-1 | Mvan_3812 | - | 3e-37 | 92.59% (81) | hypothetical protein Mvan_3812 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2724|M.gilvum_PYR-GCK ----------------MIGYVAAAAVGYVLGTKAGRRRFEQISGTYRAVT
Mvan_3812|M.vanbaalenii_PYR-1 MDPDTSTHFETSGRSPVIGYVAAAAVGYVLGTKAGRRRFEQISGTYRAVT
MSMEG_4479|M.smegmatis_MC2_155 ----------------MIRYAAVIAVGYVLGAKAGRRRYEQIASTYRAVT
TH_0724|M.thermoresistible__bu ----------------VIGYVIVLGVGYVLGSKAGRRRYEQIAGTYRAVT
MAB_1709c|M.abscessus_ATCC_199 ----------------MIRVLLVLAAGYVLGAKAGRRRYEQIVNGYQAVT
Mb2371|M.bovis_AF2122/97 ----------------MIGYVAVLGLGYVLGAKAGRRRYEQIASTYRALT
Rv2342|M.tuberculosis_H37Rv ----------------MIGYVAVLGLGYVLGAKAGRRRYEQIASTYRALT
MLBr_00834|M.leprae_Br4923 -MRQDGASRTVVGGTALIRYVIVLGLGYVLGAKAGRRRYEQIVGIYRTLT
MAV_2044|M.avium_104 ----------------MIRYVVVLGLGYVLGSKAGRRRYEQIVGTYRALT
MMAR_3649|M.marinum_M ----------------MIGYVALLGLGYVLGAKAGRRRYEQIAGTYRALT
MUL_1460|M.ulcerans_Agy99 ----------------MIGYVALLGLGYVLGAKAGRRRYEQIAGTYRALT
:* . *****:******:*** . *:::*
Mflv_2724|M.gilvum_PYR-GCK ESPAAKAVLDAGRRKIAERVSPDPAMVKLTRIDSGTEVLEPEPIVKKESR
Mvan_3812|M.vanbaalenii_PYR-1 GSPAAKAVIEAGRRKIAERVSPDPAMVKLTRIDSGTEVLEPEQMVRKNL-
MSMEG_4479|M.smegmatis_MC2_155 SNPATRAVIDAGRRKIASRVSPDPQFVTMTPIDAETSVLSVEDRTDASAK
TH_0724|M.thermoresistible__bu SSPAAKALIDAGRRKIAERVSPDPQFVKVAAIDANTTVLQPET----NGN
MAB_1709c|M.abscessus_ATCC_199 GSPATKKLIEISRRRLANSLSPDPHMVELTPIDETTMILEPEPQLDKK--
Mb2371|M.bovis_AF2122/97 GSPVARSMIEGGRRKIANRISPDAGFVTLAEIDNQTAVVQRGVERQPKTA
Rv2342|M.tuberculosis_H37Rv GSPVARSMIEGGRRKIANRISPDAGFVTLAEIDNQTAVVQRGVERQPKTA
MLBr_00834|M.leprae_Br4923 GSPMAKSMIAEGRRKVANRISPDEGFVTLAEIDNQTTVIERSAEWRENGG
MAV_2044|M.avium_104 SSPVAKSVIEGGRRKIANRISPDAGFVTLAQIDDRTAIMEREAEPADEAL
MMAR_3649|M.marinum_M GSPAAKTVIDAGRRKIANRVSPDSGFVTLTEIDTQTVVVERQPKAAP---
MUL_1460|M.ulcerans_Agy99 GSPAAKTVIDAGRRKIANRVSPDSGFVTLTEIDTQTVVVERQPKAAP---
.* :: :: .**::*. :*** :* :: ** * ::.
Mflv_2724|M.gilvum_PYR-GCK EGR----
Mvan_3812|M.vanbaalenii_PYR-1 -------
MSMEG_4479|M.smegmatis_MC2_155 SKRGW--
TH_0724|M.thermoresistible__bu ARR----
MAB_1709c|M.abscessus_ATCC_199 -------
Mb2371|M.bovis_AF2122/97 R------
Rv2342|M.tuberculosis_H37Rv R------
MLBr_00834|M.leprae_Br4923 N------
MAV_2044|M.avium_104 APSNRPR
MMAR_3649|M.marinum_M -------
MUL_1460|M.ulcerans_Agy99 -------