For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPAAIRRGEGELSVKVNTSRVIAGAFAAAAVAFPLYASLTTADTAGETIAAPRCLAWFGNQADGHCLSYS NGSGATIGTPQIGLGGGGMYINTAPLLPGTTISQPIG
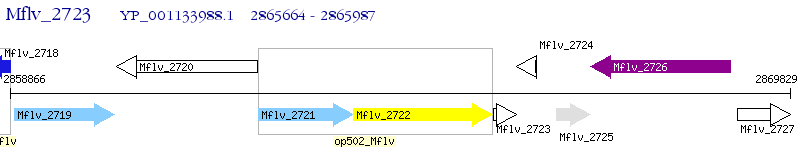
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2723 | - | - | 100% (107) | hypothetical protein Mflv_2723 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4480 | - | 3e-14 | 59.02% (61) | hypothetical protein MSMEG_4480 |
| M. thermoresistible (build 8) | TH_4379 | - | 3e-24 | 59.34% (91) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3813 | - | 2e-33 | 69.15% (94) | hypothetical protein Mvan_3813 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2723|M.gilvum_PYR-GCK ---MPAAIRRGEGELS-VKVNTSRVIAGAFAAAAVAFPLYASLTTADTAG
Mvan_3813|M.vanbaalenii_PYR-1 MAAKPSGRRRAVMLVDKIQTNKTRLIAGAIAAAAVAVPLYTSLTTVDAAP
MSMEG_4480|M.smegmatis_MC2_155 --------------------------------------------------
TH_4379|M.thermoresistible__bu --------------------MRRRLIAGALAAVAVAAPAALTAATGAAES
Mflv_2723|M.gilvum_PYR-GCK ETIAAPRCLAWFGNQADGHCLSYSNGSGATIGTPQIGLGGGG---MYINT
Mvan_3813|M.vanbaalenii_PYR-1 EVNAGPQCLAWFGNKEDGNCLSYSNGTGATIGTPQIGFGDNG---FGFYT
MSMEG_4480|M.smegmatis_MC2_155 -MTAAPACLAWFGNKEDGKCLSYSNGTPVGGGIGPVTVGAPGSGNPGFTT
TH_4379|M.thermoresistible__bu EVTAAPRCLAWFGNKEDGKCLSYSNGQPVVGGMPAIGIGGPGSSSPGIYT
*.* *******: **:******* . * : .* * : *
Mflv_2723|M.gilvum_PYR-GCK APLLPGTTISQPIG--
Mvan_3813|M.vanbaalenii_PYR-1 GPLLPGTTISQGIGGN
MSMEG_4480|M.smegmatis_MC2_155 GPLLPGTTINQGLG--
TH_4379|M.thermoresistible__bu GPLLPGTTITQPIG--
.********.* :*