For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VARDYYGLLGVSKGASDSEIKRAYRKLARELHPDVNPDEGAQARFQEISAAYEVLSDPEKRRIVDLGGDP LESAAAGGGGFSGFGGLGDVFEAFFGGGTASRGPIGRVRPGSDSLLRMRLDLAECATGVTKQVTVDTAVL CDLCHGKGTHGDSTPTSCDTCGGRGEVQTVQRSLLGQVMTSRPCPVCGGVGEVIPNPCNRCGGDGRVRAR REISVKIPAGVGDGMRVRLAAQGEVGPGGGPAGDLYVEVHEKPHPVFVRDGDDLHCTVSVPMVDAALGTT VSVDAILDGPTDITIPAGTQPGAVTTLRGHGMPHLRSGVRGDLHAHIDVVVPTRLDHEDIELLRALKERT RAAAEVRSAQSSGNGSGGGLFSKLRETFTGR
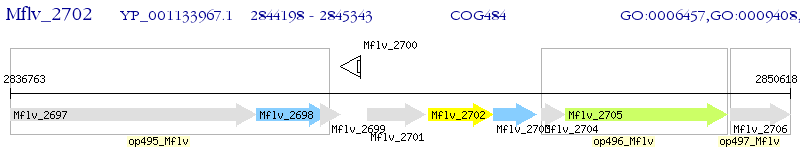
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2702 | - | - | 100% (381) | chaperone protein DnaJ |
| M. gilvum PYR-GCK | Mflv_0258 | - | 6e-68 | 39.59% (394) | chaperone protein DnaJ |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2394c | dnaJ2 | 0.0 | 82.38% (386) | chaperone protein DnaJ2 |
| M. tuberculosis H37Rv | Rv2373c | dnaJ2 | 0.0 | 82.38% (386) | chaperone protein DnaJ2 |
| M. leprae Br4923 | MLBr_00625 | dnaJ2 | 1e-177 | 79.06% (382) | chaperone protein DnaJ |
| M. abscessus ATCC 19977 | MAB_1666 | - | 1e-173 | 77.14% (385) | chaperone protein DnaJ |
| M. marinum M | MMAR_3683 | dnaJ2 | 0.0 | 82.98% (382) | chaperone protein DnaJ2 |
| M. avium 104 | MAV_2023 | dnaJ | 0.0 | 83.68% (386) | chaperone protein DnaJ |
| M. smegmatis MC2 155 | MSMEG_4504 | dnaJ | 0.0 | 86.91% (382) | chaperone protein DnaJ |
| M. thermoresistible (build 8) | TH_1931 | dnaJ2 | 0.0 | 87.79% (385) | PROBABLE CHAPERONE PROTEIN DNAJ2 |
| M. ulcerans Agy99 | MUL_3626 | dnaJ2 | 0.0 | 82.72% (382) | chaperone protein DnaJ2 |
| M. vanbaalenii PYR-1 | Mvan_3836 | - | 0.0 | 92.71% (384) | chaperone protein DnaJ |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2702|M.gilvum_PYR-GCK MARDYYGLLGVSKGASDSEIKRAYRKLARELHPDVNPDEGAQARFQEISA
Mvan_3836|M.vanbaalenii_PYR-1 MARDYYGLLGVSKGASDSEIKRAYRKLARELHPDVNPDEGAQARFQEISV
MSMEG_4504|M.smegmatis_MC2_155 MARDYYGLLGVSKGASDSEIKRAYRRLARELHPDVNPDEEAQHRFTEIQQ
TH_1931|M.thermoresistible__bu VARDYYGLLGVSKGASDTEIKRAYRRLARELHPDVNPDEEAQARFKEISV
MMAR_3683|M.marinum_M MARDYYGLLGVSKGASDAEIKRAYRKLARELHPDVNPDEAAQAKFKEISV
MUL_3626|M.ulcerans_Agy99 MARDYYGLLGVSKGASDAEIKRAYRKLARELHPDVNPDEAAQAKFKEISV
Mb2394c|M.bovis_AF2122/97 MARDYYGLLGVSKNASDADIKRAYRKLARELHPDVNPDEAAQAKFKEISV
Rv2373c|M.tuberculosis_H37Rv MARDYYGLLGVSKNASDADIKRAYRKLARELHPDVNPDEAAQAKFKEISV
MLBr_00625|M.leprae_Br4923 MARDYYGLLGVSRNASDADIKRAYRKLARELHPDINPDEAAQAKFKEISM
MAV_2023|M.avium_104 MARDYYGLLGVSRGASDAEIKRAYRKLARELHPDVNPDEAAQAKFKEISV
MAB_1666|M.abscessus_ATCC_1997 MARDYYGILGVSKGASDSELKRAYRKLARELHPDINPDEQAQARFKEVSI
:******:****:.***:::*****:********:**** ** :* *:.
Mflv_2702|M.gilvum_PYR-GCK AYEVLSDPEKRRIVDLGGDPLESAAA-GGGG-FSGFG----GLGDVFEAF
Mvan_3836|M.vanbaalenii_PYR-1 AYEVLNDPEKRRIVDLGGDPMESAAA-GGGGGFAGFG----GLGDVFEAF
MSMEG_4504|M.smegmatis_MC2_155 AYEVLSDPEKRRIVDMGGDPMESVG--GAPNGFGGFG----GLGDVFEAF
TH_1931|M.thermoresistible__bu AYEVLSDPEKRRIVDMGGDPLESPGAAGAGNGFAGFG----GLGDVFEAF
MMAR_3683|M.marinum_M AYEVLSDPEKRRIVDLGGDPLEGAGA--AANGFSGFG----GLGDVFEAF
MUL_3626|M.ulcerans_Agy99 AYEVLSDPEKRRIVDLGGDPLEGAGA--AANGFSGFG----GLGDVFEAF
Mb2394c|M.bovis_AF2122/97 AYEVLSDPDKRRIVDLGGDPLESAAA--GGNGFGGFG----GLGDVFEAF
Rv2373c|M.tuberculosis_H37Rv AYEVLSDPDKRRIVDLGGDPLESAAA--GGNGFGGFG----GLGDVFEAF
MLBr_00625|M.leprae_Br4923 AYEVLSDPEKRRIVDLGGDPMENAAA--SPSGFGGFG----GLGDVFEAF
MAV_2023|M.avium_104 AYEVLSDPEKRRIVDLGGDPLEGAAA--AGGGFGGFG----GLGDVFEAF
MAB_1666|M.abscessus_ATCC_1997 AYEVLTDPEKRRVVDLGGDPLENGGG---GNGFGGFGSGFGGLGDVFEAF
*****.**:***:**:****:*. . . *.*** *********
Mflv_2702|M.gilvum_PYR-GCK FGG----GTASRGPIGRVRPGSDSLLRMRLDLAECATGVTKQVTVDTAVL
Mvan_3836|M.vanbaalenii_PYR-1 FGG----GATSRGPVGRVRPGSDSLLRMRLDLEECATGVTKQVTVDTAVL
MSMEG_4504|M.smegmatis_MC2_155 FGG----GTTSRGPIGRVRPGSDSLLRMRLDLEECATGVTKEVTVETAVL
TH_1931|M.thermoresistible__bu FGG----GTASRGPIGRVRPGSDSLLRLQLDLEECATGVTKRVTVDTAVL
MMAR_3683|M.marinum_M FGG----GSASRGPIGRVRPGSDSLLRMRLDLEECATGVTKQVTVDTAVL
MUL_3626|M.ulcerans_Agy99 FGG----GSASRGPIGRVRPGSDSLLRMRLDLEECATGVTKQVTVDTAVL
Mb2394c|M.bovis_AF2122/97 FGGGFGGGAASRGPIGRVRPGSDSLLRMRLDLEECATGVTKQVTVDTAVL
Rv2373c|M.tuberculosis_H37Rv FGGGFGGGAASRGPIGRVRPGSDSLLRMRLDLEECATGVTKQVTVDTAVL
MLBr_00625|M.leprae_Br4923 FGG----SSASRGPIGRVRPGSDSLLPMWLDLEECATGVTKQVTVDTAVL
MAV_2023|M.avium_104 FGGGFGGGTTSRGPIGRVRPGSDSLLRMRLDLEECATGVTKQVTVDTAVL
MAB_1666|M.abscessus_ATCC_1997 FGG---SAGGSRGPRGRVQPGADSLLRMRLDLVECATGVSKQVTVDTAIL
*** . **** ***:**:**** : *** ******:*.***:**:*
Mflv_2702|M.gilvum_PYR-GCK CDLCHGKGTHGDSTPTSCDTCGGRGEVQTVQRSLLGQVMTSRPCPVCGGV
Mvan_3836|M.vanbaalenii_PYR-1 CDLCHGKGTHGDSTPVTCDTCGGRGEIQTVQRSLLGQVMTSRPCPVCGGV
MSMEG_4504|M.smegmatis_MC2_155 CDLCHGKGTNGNSSPATCDTCGGRGEIQTVQRSLLGQVMTSRPCPVCGGV
TH_1931|M.thermoresistible__bu CDLCHGKGTHGNSTPVTCDTCGGRGEIQTVQRSLLGQVMTSRPCPVCGGV
MMAR_3683|M.marinum_M CDRCQGKGTNGDSAPIPCDTCGGRGEVQTVQRSLLGQMLTSRPCPTCRGV
MUL_3626|M.ulcerans_Agy99 CDRCQGKGTNGDSASIPCDTCGGRGEVQTVQRSLLGQMLTSRPCPTCRGV
Mb2394c|M.bovis_AF2122/97 CDRCQGKGTNGDSVPIPCDTCGGRGEVQTVQRSLLGQMLTSRPCPTCRGV
Rv2373c|M.tuberculosis_H37Rv CDRCQGKGTNGDSVPIPCDTCGGRGEVQTVQRSLLGQMLTSRPCPTCRGV
MLBr_00625|M.leprae_Br4923 CDRCQGKGTNGDSAPIPCDTCGGRGEVQTVQRSLLGQMVTARPCPTCRGV
MAV_2023|M.avium_104 CDRCHGKGTNGDSAPVPCDTCGGRGEVQTVQRSLLGQVMTSRPCPTCRGV
MAB_1666|M.abscessus_ATCC_1997 CDGCAGKGTHGNSAPSACETCGGRGEVQTVQRSLLGQVLTSRPCPTCQGA
** * ****:*:* . .*:*******:**********::*:****.* *.
Mflv_2702|M.gilvum_PYR-GCK GEVIPNPCNRCGGDGRVRARREISVKIPAGVGDGMRVRLAAQGEVGPGGG
Mvan_3836|M.vanbaalenii_PYR-1 GEVIPNPCNRCSGDGRVRARREISVKIPAGVGDGMRVRLAAQGEVGPGGG
MSMEG_4504|M.smegmatis_MC2_155 GEVIPDPCHRCGGDGRVRARRDISVKIPAGVGDGMRVRLAAQGEVGPGGG
TH_1931|M.thermoresistible__bu GEVIPDPCHRCGGDGRVRARREISVKIPAGVGDGMRVRLAAQGEVGPGGG
MMAR_3683|M.marinum_M GVVIPDPCHQCMGDGRVRARREISVKIPAGVGDGMRVRLAAQGEVGPGGG
MUL_3626|M.ulcerans_Agy99 GVVIPDPCHQCMGDGRVRARREISVKIPAGVGDGMRVRLAAQGEVGPGGG
Mb2394c|M.bovis_AF2122/97 GVVIPDPCQQCMGDGRIRARREISVKIPAGVGDGMRVRLAAQGEVGPGGG
Rv2373c|M.tuberculosis_H37Rv GVVIPDPCQQCMGDGRIRARREISVKIPAGVGDGMRVRLAAQGEVGPGGG
MLBr_00625|M.leprae_Br4923 GVVIPDPCCQCVGDGRVRARREITVKIPSGVGDGMRVRLAAQGEVGPGGG
MAV_2023|M.avium_104 GVVIPDPCHQCMGDGRVRARREISVKIPAGVGDGMRVRLAAQGEVGPGGG
MAB_1666|M.abscessus_ATCC_1997 GEVITDPCHKCGGDGRVRARREITVKIPAGVGDGMRVRLAAQGEVGPGGG
* **.:** :* ****:****:*:****:*********************
Mflv_2702|M.gilvum_PYR-GCK PAGDLYVEVHEKPHPVFVRDGDDLHCTVSVPMVDAALGTTVSVDAILDGP
Mvan_3836|M.vanbaalenii_PYR-1 PAGDLYVEVHEKPHPVFVRDGDDLHCTVSVPMVDAALGTTVTVDAILDGP
MSMEG_4504|M.smegmatis_MC2_155 PAGDLYVEVHEKPHDVFVRDGDDLHCTISVPMVDAALGCTVTVDAILDGP
TH_1931|M.thermoresistible__bu PAGDLYVEVHEKPHPVFVRDGDDLHCTVSVPMVDAALGTTVVVDAILDGP
MMAR_3683|M.marinum_M PSGDLYVEVHEQAHDIFVREGDDLHCTVSVPMVDAALGVTVTLDAILDGP
MUL_3626|M.ulcerans_Agy99 PSGDLYVEVHEQAHDIFVREGDDLHCTVSVPMVDAALGVTVTLDAILDGP
Mb2394c|M.bovis_AF2122/97 PAGDLYVEVHEQAHDVFVREGDHLHCTVSVPMVDAALGVTVTVDAILDGL
Rv2373c|M.tuberculosis_H37Rv PAGDLYVEVHEQAHDVFVREGDHLHCTVSVPMVDAALGVTVTVDAILDGL
MLBr_00625|M.leprae_Br4923 PAGDLYVEVHEQAHDIFVRDGDDLHCTVSVPMVDAALGTTITVDAILDGM
MAV_2023|M.avium_104 PAGDLYVEVHEQPHDVFVREGDDLHCTVSVPMVDAALGATVTVDAILDGV
MAB_1666|M.abscessus_ATCC_1997 PAGDLYVEVHEKPHDVFIRDGDDLHFTVRVPMVEAALGTSVSVEAILDGD
*:*********:.* :*:*:**.** *: ****:**** :: ::*****
Mflv_2702|M.gilvum_PYR-GCK TDITIPAGTQPGAVTTLRGHGMPHLRSGVRGDLHAHIDVVVPTRLDHEDI
Mvan_3836|M.vanbaalenii_PYR-1 TEITIAAGTQPGAVTTLRGHGMPHLRSGVRGDLHAHIDVVVPSRLDHEDI
MSMEG_4504|M.smegmatis_MC2_155 TELTIPPGTQPGSVTTLRGHGMPHLRSGVRGDLHAHIDVVVPTRLENSDI
TH_1931|M.thermoresistible__bu IELTIPPGTQPGAVITLRGHGMPHLRSGVRGDLHAHIEVVVPTRLDPDDA
MMAR_3683|M.marinum_M SEIVIPPGTQPAAVITLRGHGMPHLRSSARGDLHVHVDVMVPVRLDHRDT
MUL_3626|M.ulcerans_Agy99 SEIVIPPGTQPAAVITLRGHGMPHLRSSARGDLHVHVDVMVPVRLDHRDT
Mb2394c|M.bovis_AF2122/97 SEITIPPGTQPGSVITLRGRGMPHLRSNTRGDLHVHVEVVVPTRLDHQDI
Rv2373c|M.tuberculosis_H37Rv SEITIPPGTQPGSVITLRGRGMPHLRSNTRGDLHVHVEVVVPTRLDHQDI
MLBr_00625|M.leprae_Br4923 SEIDISPGTQPGSVIELRGQGMPHLRSNTRGNLHVHVEVMVPTRLDQHDT
MAV_2023|M.avium_104 SEITIPPGTQPGSVITLRGHGMPHLRSGTRGDLHVHVEVVVPSRLDARDT
MAB_1666|M.abscessus_ATCC_1997 TVIKVEPGSQPGSVVTLRGKGMPHLRTGVRGNLHAHLDVVVPTRLDSKER
: : .*:**.:* ***:******:..**:**.*::*:** **: :
Mflv_2702|M.gilvum_PYR-GCK ELLRALKE-RTRAAAEVRSAQSSGNGS--GGGLFSKLRETFTGR
Mvan_3836|M.vanbaalenii_PYR-1 ELLRTFKE-RSKDTAEVRSAQG-GNGSSHGGGLFSRLRETFTGR
MSMEG_4504|M.smegmatis_MC2_155 ELLRKFKENRTRDTASVRTAQG--SAGS-GGGLFSRLRETFTGR
TH_1931|M.thermoresistible__bu ELLRKVKSNRSRDVAEVRSRLSGESSSS-GGGLFSRLRETFTGR
MMAR_3683|M.marinum_M ELLRELKNRRGRDVAEVRSSHA------AGGGLFSKLRETFSGR
MUL_3626|M.ulcerans_Agy99 ELLRELKNRRGRDVAEVRSSHA------AGGGLFSKLRETFSGR
Mb2394c|M.bovis_AF2122/97 ELLRELKGRRDREVAEVRSTHA------AAGGLFSRLRETFTGR
Rv2373c|M.tuberculosis_H37Rv ELLRELKGRRDREVAEVRSTHA------AAGGLFSRLRETFTGR
MLBr_00625|M.leprae_Br4923 ELLRELKRRRSRDVAEVRSTHA------AGGGLFSRLRETFTNR
MAV_2023|M.avium_104 ELLREFKARRGREVPEVRSTHA------GGG-LFSRLRETFTGR
MAB_1666|M.abscessus_ATCC_1997 ELLKDFRA-RNKEGAEVVWAES------ASGGVFSRLREAFTGR
***: .: * : ..* . .* :**:***:*:.*