For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDSPRCRDYAEVTEVRATEPAAVARAWETRTTRPTMRGNGRLMIVAADHPARGALSVGSRPTAMNSRTDL LDRLRAALAEPGVDGVLATADILDDLVLLGALEDKVVFSSFNRGGLAGSSFELDDRMTGATAASTAAARM NGGKMLCRIDLDDPGTVSTMAACAQAVDELAAHGLIAMLEPFLSSRVDGRVRNDLSPDAVIKSVHIGQGL GSTSAYTWMKLPVVDEMDRVMESTTMPTLLLGGDPADPEAAFASWEKALSLPSVRGLIVGRTLLYPPDDD VGAAVATAVGMIR*
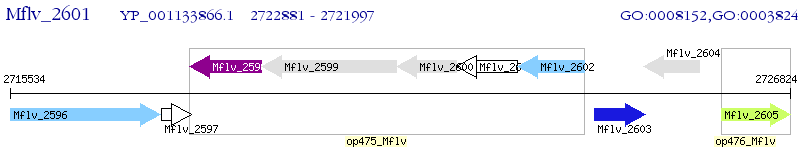
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2601 | - | - | 100% (294) | hypothetical protein Mflv_2601 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4662 | - | 1e-136 | 80.20% (293) | deoxyribose-phosphate aldolase superfamily protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4017 | - | 1e-151 | 89.80% (294) | hypothetical protein Mvan_4017 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2601|M.gilvum_PYR-GCK MSDSPRCRDYAEVTEVRATEPAAVARAWETRTTRPTMRGNGRLMIVAADH
Mvan_4017|M.vanbaalenii_PYR-1 MSDTAVCADYASLTEVRATDPGAVARAWAQRTTRPTLRGNGRLMIVAADH
MSMEG_4662|M.smegmatis_MC2_155 MSRPAVCSGHSAITDLRASEPGMIAAAWQRRTTRPTVRGDGRLMIVAADH
** .. * .:: :*::**::*. :* ** ******:**:**********
Mflv_2601|M.gilvum_PYR-GCK PARGALSVGSRPTAMNSRTDLLDRLRAALAEPGVDGVLATADILDDLVLL
Mvan_4017|M.vanbaalenii_PYR-1 PARGALAVGGRPTAMNSRTDLLDRLQAALADPGVDGVLATSDILDDLLLL
MSMEG_4662|M.smegmatis_MC2_155 PARGALAVGSRPTAMNSRTDLLDRLCTALADPGVDGVLATADILDDLLLL
******:**.*************** :***:*********:******:**
Mflv_2601|M.gilvum_PYR-GCK GALEDKVVFSSFNRGGLAGSSFELDDRMTGATAASTAAARMNGGKMLCRI
Mvan_4017|M.vanbaalenii_PYR-1 GALEDKVVFSSFNRGGLAGSSFELDDRMTGATAASTAAARMNGGKMLCRI
MSMEG_4662|M.smegmatis_MC2_155 GALEDKVVFSSFNRGGLSGSVFEMDDRMTGATAASTAAARMNGGKMLCRI
*****************:** **:**************************
Mflv_2601|M.gilvum_PYR-GCK DLDDPGTVSTMAACAQAVDELAAHGLIAMLEPFLSSRVDGRVRNDLSPDA
Mvan_4017|M.vanbaalenii_PYR-1 DLDDPGTVSTLAACAQAVDELAAHGLIAMLEPFMSVRVDGKVRNDLTPDA
MSMEG_4662|M.smegmatis_MC2_155 DLDDPGTVATLASCAQAVDALASHGLVAMLEPFLSRRVDGKVRNDLTPDA
********:*:*:****** **:***:******:* ****:*****:***
Mflv_2601|M.gilvum_PYR-GCK VIKSVHIGQGLGSTSAYTWMKLPVVDEMDRVMESTTMPTLLLGGDPADPE
Mvan_4017|M.vanbaalenii_PYR-1 VIKSVHIGQGLGSTSAYTWMKLPVVDEMERVMESTTMPTLLLGGDPTDPE
MSMEG_4662|M.smegmatis_MC2_155 VIKSVHISQALGSTSAYTWLKLPVVDEMERVMDATTMPTLLLGGDPTDPD
*******.*.*********:********:***::************:**:
Mflv_2601|M.gilvum_PYR-GCK AAFASWEKALSLPSVRGLIVGRTLLYPPDDDVGAAVATAVGMIR
Mvan_4017|M.vanbaalenii_PYR-1 EAFSSWEKALSLPSVRGLIVGRTLLYPPDDDVSSAVATAVAMVR
MSMEG_4662|M.smegmatis_MC2_155 EAFASWEKALALPSVRGLIAGRSLLYPADDDVRSAISTAVKLVH
**:******:********.**:****.**** :*::*** :::